YB1 regulates miR-205/200b-ZEB1 axis by inhibiting microRNA maturation in hepatocellular carcinoma
Abstract
Background
Y-box binding protein 1 (YB1 or YBX1) plays a critical role in tumorigenesis and cancer progression. However, whether YB1 affects malignant transformation by modulating non-coding RNAs remains largely unknown. This study aimed to investigate the relationship between YB1 and microRNAs and reveal the underlying mechanism by which YB1 impacts on tumor malignancy via miRNAs-mediated regulatory network.
Methods
The biological functions of YB1 in hepatocellular carcinoma (HCC) cells were investigated by cell proliferation, wound healing, and transwell invasion assays. The miRNAs dysregulated by YB1 were screened by microarray analysis in HCC cell lines. The regulation of YB1 on miR-205 and miR-200b was determined by quantitative real-time PCR, dual-luciferase reporter assay, RNA immunoprecipitation, and pull-down assay. The relationships of YB1, DGCR8, Dicer, TUT4, and TUT1 were identified by pull-down and coimmunoprecipitation experiments. The cellular co-localization of YB1, DGCR8, and Dicer were detected by immunofluorescent staining. The in vivo effect of YB1 on tumor metastasis was determined by injecting MHCC97H cells transduced with YB1 shRNA or shControl via the tail vein in nude BALB/c mice. The expression levels of epithelial to mesenchymal transition markers were detected by immunoblotting and immunohistochemistry assays.
Results
YB1 promoted HCC cell migration and tumor metastasis by regulating miR-205/200b‒ZEB1 axis partially in a Snail-independent manner. YB1 suppressed miR-205 and miR-200b maturation by interacting with the microprocessors DGCR8 and Dicer as well as TUT4 and TUT1 via the conserved cold shock domain. Subsequently, the downregulation of miR-205 and miR-200b enhanced ZEB1 expression, thus leading to increased cell migration and invasion. Furthermore, statistical analyses on gene expression data from HCC and normal liver tissues showed that YB1 expression was positively associated with ZEB1 expression and remarkably correlated with clinical prognosis.
Conclusion
This study reveals a previously undescribed mechanism by which YB1 promotes cancer progression by regulating the miR-205/200b‒ZEB1 axis in HCC cells. Furthermore, these results highlight that YB1 may play biological functions via miRNAs-mediated gene regulation, and it can serve as a potential therapeutic target in human cancers.
Abbreviations
-
- 3′UTR
-
- 3′ untranslated region
-
- Ago2
-
- argonaute-2
-
- ATCC
-
- American Type Culture Collection
-
- ATP
-
- adenine triphosphate
-
- BEGM
-
- bronchial epithelial cell growth medium
-
- CSD
-
- cold shock domain
-
- CTCC
-
- Committee on Type Culture Collection of the Chinese Academy of Science
-
- CTD
-
- C-terminal domain
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- EGF
-
- epidermal growth factor
-
- EMT
-
- epithelial to mesenchymal transition
-
- FBS
-
- fetal bovine serum
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- miRNA
-
- microRNA
-
- ORF
-
- open reading frame
-
- pre-miRNA
-
- precursor miRNA
-
- pri-miRNA
-
- primary transcripts of miRNA
-
- qPCR
-
- quantitative real-time PCR
-
- SD
-
- standard deviation
-
- SFB
-
- s-protein, flag tag and streptavidin-binding peptide
-
- Shad1
-
- short RNA antisense to Dicer1
-
- shRNA
-
- short hairpin RNA
-
- siRNA
-
- small interfering RNA
-
- TCGA
-
- The Cancer Genome Atlas
-
- TUT
-
- terminal uridylyltransferase
-
- UTP
-
- uridine triphosphate
-
- YB1
-
- Y-box binding protein 1
-
- ZEB1
-
- zinc-finger E-box binding homeobox 1
1 INTRODUCTION
Y-box binding protein 1 (YB1 or YBX1), a DNA/RNA binding protein, is involved in multiple cellular processes, such as DNA transcription, pre-mRNA splicing, cap-dependent mRNA translation, drug-resistant, and DNA reparation [1-3]. Numerous studies have shown that YB1 exhibits critical roles in tumorigenesis and cancer progression [4-7]. For instance, YB1 can induce epithelial to mesenchymal transition (EMT) and invasion by the translational activation of Snail in breast cancer [8-10]. Recently, much attention has been focused on the relationships between YB1 and functional non-coding RNAs in various cancers [11-14]. Wu et al. [5] found that YB1 promoted cell proliferation by repressing miR-29 expression in glioblastoma multiforme. Liu et al. [14] reported that YB1 was associated with nonpolyadenylated short RNA antisense to Dicer1 (Shad1) and contributed to prostate cancer growth. YB1 may also play regulatory functions on non-coding RNAs in tumorigenesis and malignant transformation. However, the mechanisms by which YB1 modulates aberrant microRNAs (miRNAs) expression in cancers are not completely understood.
miRNAs refer to a class of small non-coding RNAs with ∼22 bp in length. Accumulating evidences have shown that miRNAs have critical functions in tumorigenesis and malignancy by regulating target mRNAs in human cancers [15-21]. The miR-200 family members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) and miR-205 are well known to suppress EMT and invasion through the downregulation of ZEB1 and SIP1 expression in cancer cells [15, 20, 22]. However, the reasons leading to the dysregulation of miRNAs expression in cancers remain largely unknown. Mechanistically, miRNAs are processed in multiple steps by microprocessors. First, the primary transcripts of miRNAs (pri-miRNAs) are cleaved by the microprocessor complexes of Drosha and DGCR8 into precursor miRNAs (pre-miRNAs) in the nucleus [23-28]. Next, pre-miRNAs are exported by Exportin-5 to the cytoplasm and further processed by a Dicer complex into a miRNA duplex [29, 30]. Finally, argonaute-2 (Ago2) corporates with a RNA-induced silencing complex and divides the miRNA duplex into a single-stranded miRNA, which further suppresses the target mRNAs by complementally binding to their 3′ untranslated regions (3′UTR) [31, 32]. It has been shown that factors associated with the above microprocessors, such as DDX1, Hippo, p68 and p72, could influence the expression and functions of miRNAs [33-37].
Recently, the terminal uridylyl transferases (TUTases or TUTs) have been found to contribute to miRNAs biogenesis by adding uridine triphosphate (UTP) instead of adenine triphosphate (ATP) to the terminal of pre-miRNAs. TUT4 (ZCCHC11/PAPD2/Hs3) was identified to interact with Lin28 and suppress let-7 miRNA biogenesis in embryonic stem cells, leading to pre-let-7 decayed by exonucleases [38-42]. TUT7 (ZCCHC6/PAPD6/Hs2) can recognize and uridylate pre-miRNAs with different overhangs in the absence of Lin28 [43]. TUT1 (PAPD2/Hs5) has been reported to affect U6 small nuclear RNA recycling by terminal uridylation [44, 45]. Since YB1 contains the highly conserved cold shock domain (CSD) with Lin28, it is unclear whether YB1 in combination with TUTs plays regulatory roles on non-coding RNAs in human cancers.
In this study, we investigate the relationship between YB1 and microRNAs, to figure out the regulatory mechanism how YB1 attributes to microRNA processing and explore whether YB1 plays critical roles in cancer malignancy via miRNAs-mediated pathway.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
Cell lines (SMMC7721, BEL7402, MHCC97H, HCCLM3, Huh7, HB611, and HEK293T) were purchased from the cell bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences (CTCC, Shanghai, China). Normal liver cell line (THLE2) was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cell lines SMMC7721, HCCLM3, Huh7, HB611, and HEK293T were cultured in Dulbecco's modified Eagle's medium (DMEM; GIBCO, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; GIBCO, Carlsbad, CA, USA) and 1% Penicillin-Streptomycin (P/S; GIBCO, Carlsbad, CA, USA). The cell lines MHCC97H and BEL7402 were cultured in RPMI 1640 medium (GIBCO, Carlsbad, CA, USA) supplemented with 10% FBS and 1% P/S. THLE2 cell line was cultured in bronchial epithelial cell growth medium (BEGM; Lonza/Clonetics Corporation, Walkersville, MD, USA) supplemented with 200 ng/mL epidermal growth factor (EGF; Thermo Fisher Scientific, Waltham, MA, USA), 500 ng/mL hydrocortisone (MedChemExpress, Monmouth Junction, NJ, USA), 100 ng/mL choleratoxin (MedChemExpress, Monmouth Junction, NJ, USA), 10 μg/mL insulin (MedChemExpress, Monmouth Junction, NJ, USA), 10% FBS and 1% P/S. All cells were grown in a humidified incubator at 37℃ in 5% CO2.
2.2 Transfection of expression plasmid, short hairpin RNA (shRNA) and small interfering RNA (siRNA)
For stable transfection of shRNAs, the cell lines were transfected by the lentiviral transduction method, as previously described [46]. Briefly, virus particles were produced by cotransfecting pCMV-VSV-G, pCMVΔ8.2 and the shRNA- or ORF-containing vector into HEK293T cells. The virus was harvested 48 h after transfection and then added to the pre-seeded target cell lines. The stable cell lines were selected using 2 μg/mL puromycin (Thermo Fisher Scientific, Waltham, MA, USA) or 10 μg/mL blasticidin (Thermo Fisher Scientific, Waltham, MA, USA) 24 h after virus infection.
For transient transfection of siRNAs or miRNA inhibitors, 5×105 cells per well were pre-coated into a six-well plate, then plasmids, siRNAs, or miRNA inhibitors were transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The transfected cells were harvested after 24 h to 48 h and were used for analyses.
YB1 open reading frame (ORF) fragments were obtained from pCMV-YB1-myc vector (GeneCopoeia, Wuhan, China). Then, YB1 ORFs were subcloned into pLOC lentivirus vector (Invitrogen, Carlsbad, CA, USA) and SFB (S-protein, Flag tag and streptavidin-binding peptide)-tagged expression vector. Similarly, the truncated YB1 constructs were produced by subcloning the truncated YB1 domains into the SFB-tagged expression vector. The pre-miR-205 and pre-miR-200b expressing plasmids were purchased from the GeneCopoeia company (Wuhan, China). Flag-tagged DGCR8, Flag-tagged Dicer, Flag-tagged TUT4, and Flag-tagged TUT1 plasmids were kindly provided by Dr. V. Narry Kim (School of the Biological Sciences, Seoul National University, Korea). The human YB1 shRNA plasmids were purchased from Sigma Company (St. Louis, MO, USA). shRNA sequences used in this study were as follows: 5′-CCGGCCAGTTCAAGGCAGTAAATATCTCGAGATATTTACTGCCTTGAACTGGTTTTTG-3′ designated for shYB1_4 (Catalog No. TRCN0000315307); 5′-CCGGAGCAGACCGTAACCATTATAGCTCGAGCTATAATGGTTACGGTCTGCTTTTTTG-3′ designated for shYB1_5 (Catalog No. TRCN0000315309). Snail siRNA (si-Snail) and negative control (si-NC) were obtained from GenePharma Company (Shanghai, China). siRNA sequences used in this study were as follows: 5′-GCCUUCAACUGCAAAUACUTTAGUAUUUGCAGUUGAAGGCTT-3′ for si-Snail; 5′-UUCUCCGAACGUGUCACGUTT-3′ for si-NC. Snail shRNA was produced by cloning Snail shRNA sequence into pLKO.1 lentiviral vector. Snail shRNA sequence: 5′-CCGGCGCTTTGAGCTACAGGACAAACTCGAGTTTGTCCTGTAGCTCAAAGCGTTTTT-3′.
2.3 Cell proliferation, migration, and invasion assays
Cell proliferation assays were performed in triplicate using the crystal violet staining method, as previously described [46]. Briefly, 5×103 cells per well were seeded in a 6-well plate and incubated for an indicated period of time. Next, the cells were fixed with 10% methanol and stained with 0.1% crystal violet. Each well was washed three times with PBS and destained with 10% acetic acid. Finally, the number of viable cells was quantified by detecting the absorbance of the crystal violet solution at 590 nm.
The cell migration ability was detected by wound healing assay. Briefly, 1×106 cells were seeded in 6-well plates and serum-starved overnight. On the next day, the cells reached approximately 98% confluency and were cultured in serum-free medium supplemented with 4 μg/mL Mitomycin C (MedChemExpress, Shanghai, China) for 6 h to limit cell proliferation. Next, the cells were scratched with a sterile 10 μL pipette tip, and incubated in serum-free media, after cell debris were removed with PBS. The closure of the gap was photographed at an indicated period of time and measured with Image J software. The wound closure was calculated as below: wound closure = (area of gap [0 h] – area of gap [24/48 h])/area of gap (0 h).
The cell invasion ability was assayed using the Transwell plate coated with Matrigel (BD Biosciences., San Jose, CA, USA) according to the manufacture's instructions. Briefly, the cells were serum-starved overnight before plating. On the next day, 2×105 cells per well were seeded in the Transwell plate and incubated for 30 h. Finally, the invasive cells were fixed with 10% methanol and stained with 0.1% crystal violet for 1 h. Each well was washed with PBS thrice, then photographed at three random fields. The invasive cells were counted under an inverted phase-contrast microscope (Leica Microsystems, Wetzlar, Germany).
2.4 Microarray analysis and quantitative real-time PCR (qPCR)
Total RNA, inclusive of small RNAs, was isolated using RNAiso reagent (Takara, Dalian, China), and then reversely transcribed into cDNA using the All-in-One miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia, Wuhan, China). MicroRNA array assays were performed using miProfile HCC miRNA qPCR arrays (GeneCopoeia, Wuhan, China) according to the manufacturer's instructions, which profiled the expression of 168 specific miRNAs associated with HCCs. Individual miRNA expression was further analyzed by qPCR with All-in-One miRNA qPCR reagents (GeneCopoeia, Wuhan, China) on a CFX96 instrument (Bio-Rad Laboratories, Hercules, CA, USA). The specific qPCR primers were listed in Supplementary Table S1. All relative gene expression levels were measured by at least three biological replicates for different experimental groups and with three technical replicates per biological replicate. The relative expression levels of miRNAs were calculated using the 2-△△Ct method after normalization to the U6 expression.
2.5 Luciferase reporter assay
The luciferase reporter plasmids with ZEB1 or SIP1 3′UTRs were constructed by amplifying the gene fragments from THLE2 cDNA using the primers described previously [15], then subcloned into a pEZX-FR02 vector with firefly luciferase (hLuc) and renilla luciferase (Rluc) genes (GeneCopoeia, Wuhan, China). The luciferase activity assays were performed by cotransfecting ZEB1 or SIP1 3′UTR luciferase reporter plasmid, pre-miRNA expressing plasmid, and pCMV-YB1-myc plasmid into HEK293T cells. The luciferase reporter activity was measured 48 h after transfection using the Luc-Pair Duo-Luciferase HS Assay Kit according to the manufacturer's instructions (GeneCopoeia, Wuhan, China).
2.6 Pull-down and coimmunoprecipitation assay
The pull-down experiments were performed by cotransfecting SFB-tagged YB1 constructs with Flag-tagged DGCR8 or Flag-tagged Dicer plasmid into HEK293T cells. The cells were lysed 48 h after transfection in NETN buffer (200 mmol/L Tris-HCl [pH 8.0], 100 mmol/L NaCl, 0.05% NP-40 and 1 mmol/L EDTA) with protease and phosphatase inhibitors (Sigma, St Louis, MO, USA). The cell lysates were pulled down by anti-Flag Magnetic beads (Sigma, St Louis, MO, USA) or S-protein beads (Sigma, St Louis, MO, USA) as previously described [46]. The RNA-independent immunoprecipitation was performed by adding RNase A (1 μg/μL; Takara, Dalian, China) in cell lysates. The endogenous immunoprecipitation assays were performed with the primary antibody against YB1 (Cell Signaling Technology, 9744), DGCR8 (Proteintech, 10996-1-AP), Dicer (Proteintech, 20567-1-AP), and TUT4 (Proteintech, 18980-1-AP). The uncropped images of immunoblots were summarized in Supplementary materials.
2.7 Biotin pull-down and RNA immunoprecipitation assay
The pre-miRNA sequences were amplified from pre-miR-205 and pre-miR-200b expressing plasmids (GeneCopoeia, Wuhan, China) respectively, then subcloned into the downstream of SP6 or T7 promoter in pEASY-T3 vector (TransGen Biotech, Beijing, China). The biotin pull-down assays were performed as described previously [13]. Briefly, pre-miR-205 and pre-miR-200b were transcribed in vitro and biotin-labeled with MEGAscript T7 Kit containing biotin-uridine triphosphate (Thermo Scientific, Waltham, MA, USA), then purified with the MEGAclear Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Next, the biotin-labeled pre-miRNAs were incubated with the whole cell lysates. Then, the streptavidin beads were used to pull down biotin-labeled transcripts and coprecipitated proteins. Finally, the RNA affinity proteins were detected by immunoblotting assays.
RNA immunoprecipitation [47] was performed by cotransfecting SFB-tagged plasmid and pre-miRNA expressing plasmid into HEK293T cells. After 48 h, the cells were lysed in NETN buffer with protease and phosphatase inhibitors (Sigma, St Louis, MO, USA) and RNase inhibitor (1 U/μL; Takara, Dalian, China). The supernatant of cell lysates were incubated with S-protein beads on a rotating wheel for 6 h at 4℃. Next, the YB1-coprecipitated RNAs were purified with S-protein beads by centrifugation at 3000 rpm for 3 min at 4℃. After washing the beads three times with NETN buffer, the YB1-coprecipitated RNAs were extracted with RNAiso reagent (Takara, Dalian, China) and detected by qPCR analysis.
2.8 Immunoblotting assay
The immunoblotting assays were performed using standard methods. The proteins from cells were lysed in RIPA buffer with protease and phosphatase inhibitors (Sigma, St Louis, MO, USA), and harvested by centrifugation at 14000 rpm for 15 min. The protein samples from tumors were homogenized with glass beads and then lysed in RIPA buffer with protease and phosphatase inhibitors. The independent fresh HCC and adjacent non-tumor tissues were obtained from the patients during the surgical resection. All patients had signed informed consents and agreed to donate the cancer tissues for research. Ethical approval for the project was provided by the First Affiliated Hospital of Dalian Medical University Clinical Experiment Ethics Committee. The following antibodies were used: antibodies to YB1 (1:1000; 9744; Cell Signaling Technology), E-Cadherin (1:1000; 14472; Cell Signaling Technology), N-Cadherin (1:2000; 22018-1-AP; Proteintech), ZEB1 (1:1000; 21544-1-AP; Proteintech), Snail (1:1000; 3879; Cell Signaling Technology), Vinculin (1:2000; V4505; Sigma), GAPDH (1:2000; 60004-1-Ig; Proteintech), Smad3 (1:1000; 9523; Cell Signaling Technology), Akt1 (1:1000; 4691; Cell Signaling Technology), DGCR8 (1:1000; 10996-1-AP; Proteintech), Dicer (1:1000; 20567-1-AP; Proteintech), TUT4/ZCCHC11 (1:800; 18980-1-AP; Proteintech), β-Tubulin (1:1000; 10068-1-AP; Proteintech), Lamin B1 (1:1000; 12987-1-AP; Proteintech), Flag/DDDK tag (1:1000; 20543-1-AP; Proteintech), Myc-tag (1:2000; 60003-2-Ig; Proteintech) and tGFP (1:2000; TA150041; Origene). The uncropped images of immunoblots were summarized in Supplementary materials.
2.9 Immunofluorescent and immunohistochemistry assay
The endogenous protein localization was determined by immunofluorescence staining. Briefly, the cells were fixed with 4% formaldehyde, then permeated with 0.1% Triton-X-100 and blocked with 5% bovine serum albumin, and finally incubated with primary antibodies to YB1 (1:50; 4202; Cell Signaling Technology), DGCR8 (1:50; 10996-1-AP; Proteintech) and Dicer (1:50; 20567-1-AP; Proteintech) respectively at 4℃ overnight. On the next day, the cells were washed with TBST buffer (Sigma, St Louis, MO, USA) and then incubated with Alexa Fluor 488/568-conjugated anti-IgG (Thermo Fisher Scientific, Waltham, MA, USA). The nuclei were visualized using DAPI (Beyotime, Shanghai, China). Digital photographs of fluorescence were acquired with Olympus FV1000 MPE confocal laser scanning microscope (Olympus, Tokyo, Japan).
Immunohistochemistry assays were performed on formalin-fixed, paraffin-embedded lung tissues after deparaffinization and antigen retrieval. Briefly, the formalin-fixed paraffin-embedded (FFPE) lung tumor tissues were cut into 4-μm-thick slices and pretreated with Tris/EDTA buffer (pH9.0) for heat-induced epitope retrieval. Next, tissue slides were blocked with BSA and incubated with primary antibodies to YB1 (1:50; 8475; Cell Signaling Technology), ZEB1 (1:2000; 21544-1-AP; Proteintech), E-Cadherin (1:200; 14472; Cell Signaling Technology), and N-Cadherin (1:2000; 22018-1-AP; Proteintech) respectively. Finally, immunohistochemical analyses of FFPE lung tumor tissues were performed using the product from Proteintech Company (ProteinTech Group, Chicago, IL, USA) according to the manufacture's instructions.
2.10 Cytoplasmic and nuclear fractionation
Protein samples of cytoplasmic and nuclear fractions of HCC cell were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Briefly, 2×106 fresh cells were harvested for nucleocytoplasmic fractionation and followed by immunoblotting assays. Lamin B1 was used as a nuclear fraction marker. GAPDH and β-Tubulin were used as cytoplasmic markers.
2.11 Lung metastasis assay
Nude BALB/c mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Led (Beijing, China) and fed in the specific pathogen-free environments at the Institute of Genome Engineered Animal Models for Human Disease of Dalian Medical University. All animal experiments were approved by Dalian Medical University Research Ethics Committee. 1×106 MHCC97H cells stably expressing YB1 shRNA or shControl were injected into six-week-old BALB/c male nude mice (n = 10, for each experiment group) via tail vein injection. About 8 weeks after injection, mice were euthanized when they met the institutional euthanasia criteria for overall health condition. The number of nodules derived from mice lung tissues was calculated and used for immunohistochemistry and immunoblotting analyses. The expression levels of miR-205 and miR-200b were detected by qPCR analysis.
2.12 Open-source data acquisition and analysis
We obtained normalized RNA-seq data as well as the clinical information of HCC patients from TCGA [48, 49] using the GDC download function of the TCGA biolinks package [50]. The difference between YB1 expression in matched tumor and normal tissues was tested by Wilcox-test (paired, two-tailed). We also obtained the disease stage annotations from the clinical information (the disease stage recorded for each patient in the TCGA HCC samples), and the YB1 expression difference across stage I to stage III was examined by Wilcox-test (unpaired, two-tailed). The patients' survival curves were analyzed based on the Kaplan-Meier estimator. In addition, we downloaded the RNA-level expression matrix from GTEx website (https://gtexportal.org/home/datasets, version: GTEx analysis V7, data name: GTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_tpm.gct.gz) [51], extracted the expression levels of YB1 and ZEB1 in liver tissues, and evaluated their correlation by Spearman's test. All computational codes are available upon request.
2.13 Statistical analysis
All experimental data were represented as the mean ± standard deviation (SD) from at least three independent biological experiments. Statistical analysis was performed using Student's t-test (unpaired, two-tailed) to compare two groups of independent samples, unless otherwise indicated. Statistical significance was indicated as follows: n.s., no significant; *, P < 0.05; **, P < 0.001. The sample size for human patient tissues was estimated by the sample size calculator (https://clincalc.com/stats/samplesize.aspx) where the mean expression level of tumor tissues was set as 1 ± 0.3 (group 1) while the mean level for normal tissues was set as 0.5 (group 2), and the minimum sample size was 6 for each group.
3 RESULTS
3.1 YB1 enhances migration and invasion in HCC cells
YB1 is an oncogenic factor that plays critical roles in tumorigenesis and cancer progression in various cancers [5, 7, 10]. To investigate the biological functions of YB1 in HCC cells, we generated YB1 over-expressed and knockdown cell lines based on endogenous YB1 expression in a panel of HCC cells (Supplementary Figure S1A). Notably, YB1 knockdown decreased the N-cadherin and ZEB1 expression but increased E-cadherin expression in HCCLM3 and MHCC97H cells (Figure 1A). In contrast, YB1 overexpression remarkably reduced the E-cadherin expression but increased the N-cadherin and ZEB1 expression in SMMC7721 and BEL7402 cells (Figure 1B).
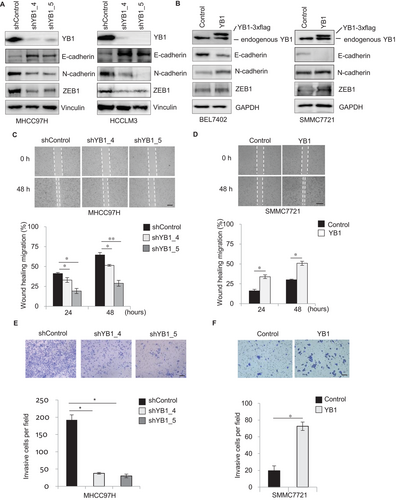
Next, we performed both loss-of-function and gain-of-function analyses of YB1 in HCC cells. YB1 knockdown by two independent shRNAs decreased the wound healing migration and cell invasion in MHCC97H and HCCLM3 cells (Figures 1C and 1E; Supplementary Figure S1B and D). In contrast, the overexpression of YB1 significantly enhanced cell migration and invasion in SMMC7721 and BEL7402 cells (Figures 1D and 1F; Supplementary Figure S1C and E). In addition, we observed that YB1 knockdown significantly reduced cell proliferation in MHCC97H and HCCLM3 cells. However, YB1 overexpression also inhibited cell growth in BEL7402 and SMMC7721 cells (Supplementary Figure S1F and G). Some studies have reported that overexpression of YB1 could promote cancer cell growth in glioblastoma multiforme [5] and breast cancer cell lines [7], but other studies have shown that enforced YB1 expression reduces cell proliferation by inhibiting cap-dependent translation of growth-related mRNAs in breast cancers [2, 10]. These reports reveal that YB1 regulates cancer cell proliferation with very complicated mechanisms [12]. Collectively, this study indicates that YB1 enhances cell migration and invasion in HCC cells.
3.2 YB1 regulates the miR-205/200b-ZEB1 axis by inhibiting miRNA expression
As the homolog of YB1, Lin28 has been reported to play biological functions by repressing let-7 miRNA biogenesis in cancers [41, 52], leading us to wonder whether YB1 could also promote cancer malignancy through the modulation of miRNAs. To identify miRNAs dysregulated by YB1 in cancer progression, we performed a miRNA qPCR array analysis in YB1 stably overexpressing HCC cells. The expression levels of 36 cancer-associated miRNAs were significantly altered in YB1 overexpressed SMMC7721 cells (Supplementary Figure S2A, cutoff > 1.5). Among these miRNAs, the levels of miR-205 and miR-200b, the well-known EMT inhibitors [15], were significantly decreased in YB1 over-expressed SMMC7721 cells. To further confirm whether YB1 could regulate these miRNAs expression, we performed qPCR analysis individually in YB1 over-expressed and knockdown HCC cells, respectively (Supplementary Figure S2B-D). As expected, the expression levels of miR-205 and miR-200b were significantly increased in YB1 knockdown MHCC97H and HCCLM3 cells, whereas they were dramatically decreased in YB1 over-expressed SMMC7721 and BEL7402 cells (Figures 2A and 2B), which were consistent with the array results. Our data indicate that YB1 could inhibit miR-205 and miR-200b expression in HCC cells.
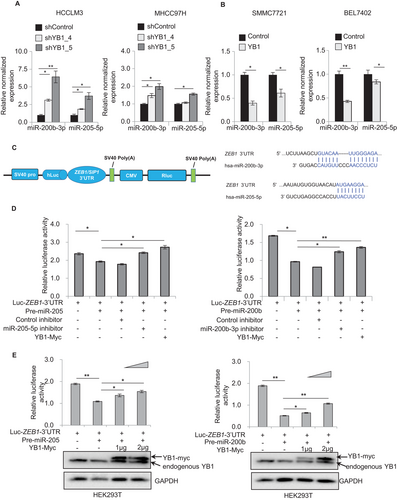
Since miR-205 and miR-200b are known to prevent EMT and cancer metastasis by suppressing ZEB1 and SIP1 expression and complementally binding to the 3′UTR of their mRNAs [15, 20, 22], we attempted to investigate whether YB1 increases ZEB1 or SIP1 expression through the downregulation of miR-205/200b using dual-luciferase reporter assays. As expected, the ZEB1 3′UTR luciferase activities were remarkably reduced by transfecting pre-miR-205 and pre-miR-200b, but were recovered by cotransfecting miR-205 or miR-200b inhibitors as well as YB1 expressing plasmid in HEK293T cells (Figures 2C and 2D). In addition, the ZEB1 and SIP1 luciferase activities were significantly increased in a YB1 dose-dependent manner (Figure 2E, Supplementary Figure S2E and F). These results suggest that YB1 could increase ZEB1 or SIP1 expression by downregulating miR-205 and miR-200b expression in HCC cells.
3.3 YB1 binds to pre-miR205/200b and inhibits miRNA maturation
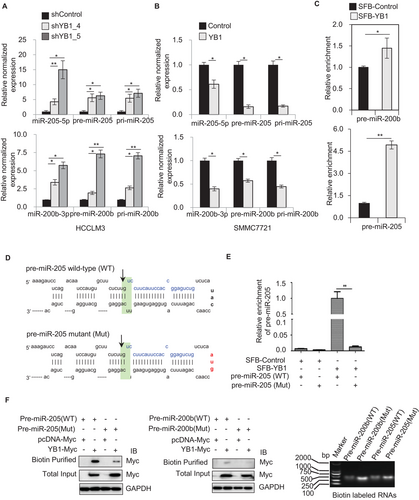
To define the molecular mechanism of YB1 regulation on miR-205/200b expression, we analyzed the expression of miR-205/200b transcripts during miRNAs processing. Knockdown of YB1 significantly increased the levels of pri-, pre- and mature miR-205 or miR-200b in both HCCLM3 and MHCC97H cells (Figure 3A, Supplementary Figure S3A). In contrast, the overexpression of YB1 remarkably decreased their expression levels in SMMC7721 and BEL7402 cells (Figure 3B, Supplementary Figure S3B). RNA immunoprecipitation assays and qPCR analysis indicated that SFB-tagged YB1 could enrich more amounts of pre-miR-205 and pre-miR-200b compared to the control in HEK293T cells (Figure 3C). To further investigate whether YB1 especially could bind to the loop region of pre-miRNAs, we produced the mutant constructs of pre-miR-205 and pre-miR-200b at the loop sequence (Figure 3D, Supplementary Figure S3C). Notably, either the mutant pre-miR-205 or pre-miR-200b enriched by SFB-tagged YB1 was significantly decreased compared to their wild-type counterparts in HEK293T cells (Figure 3E, Supplementary Figure S3D). Additionally, RNA affinity purification in vitro further confirmed that the biotin-labeled wild-type pre-miR-205 or pre-miR-200b could pull down more YB1 protein than their mutant counterparts (Figure 3F, Supplementary Figure S3E). Collectively, these results suggest that YB1 could repress the miR-205/200b maturation by binding to the loop region of pre-miRNAs.
3.4 YB1 suppresses miR-205/200b maturation by interacting with DGCR8 and Dicer
In general, miRNA maturations are processed by the microprocessor complexes of Drosha/DGCR8 in the nuclear and by Dicer in the cytoplasm, respectively [23, 26]. To investigate whether YB1 could physically interact with the main microprocessors, we first identified the cellular localization of YB1 and the microprocessors with immunofluorescent staining and cytoplasm/nuclear fractionation assay. The results indicated that YB1 co-localized with DGCR8 and Dicer respectively in HCC cells (Figure 4A, Supplementary Figure S4A and B). Pull-down experiments showed that exogenous YB1 could interact with DGCR8 or Dicer in HEK293T cells (Supplementary Figure S4C). Coimmunoprecipitation assays further confirmed that endogenous YB1 could pull down DGCR8 or Dicer, respectively, in MHCC97H, BEL7402, and HCCLM3 cells, and vice versa (Figure 4B, Supplementary Figure S4D). Consistent with previous studies [53-55], two YB1 interacting proteins Smad3 and Akt1 were also detectable in YB1 immunoprecipitates (Figure 4B).
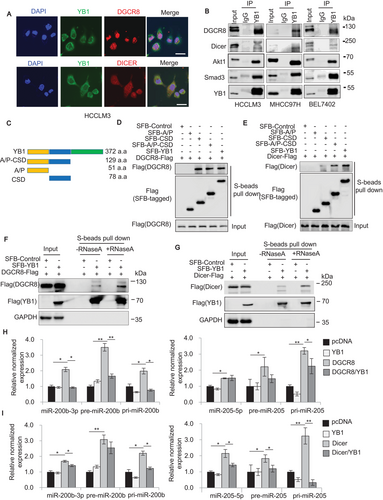
Previous studies have reported that YB1 contains three domains, including alanine and proline-rich domain (A/P), CSD, and C-terminal domain (CTD) [12, 56]. Here, we found that the conserved CSD medicated the interactions of YB1 with DGCR8 or Dicer (Figure 4C–E). To clarify whether the interactions require RNAs, we treated the cell lysates with RNase A and then carried out pull-down experiments again. The results indicated that RNase A treatment slightly affected the interactions of YB1 with DGCR8 or Dicer (Figures 4F and 4G, Supplementary Figure S4E). Furthermore, to investigate whether YB1 influences the miR-205/200b processing by microprocessors DGCR8 or Dicer, we examined the expression of miR-205 and miR-200b transcripts by qPCR analysis in HEK293T cells cotransfected with YB1 and DGCR8 or Dicer. Our data demonstrated that either DGCR8 or Dicer overexpression significantly increased the levels of pri-, pre- and mature miR-205 or miR-200b compared to the control, whereas the increased expression levels were remarkably inhibited by cotransfecting YB1 with DGCR8 or Dicer (Figures 4H and 4I, Supplementary Figure S4F). Collectively, these results suggest that YB1 may restrain miR-205/200b maturation by interacting with DGCR8 and Dicer.
3.5 YB1 inhibits miR-205/200b expression in a Snail-independent manner
YB1 induces EMT and tumor metastasis by the translational activation of the EMT inducer Snail in breast cancers [10]. Recently, another study has reported that Snail regulates miRNAs expression in the early step of metastasis in HT29 colon cancer cells [57]. To explore whether YB1 could regulate miR-205/200b expression through Snail activation, we knocked down Snail in HCC cell lines which stably expressing YB1 shRNA and shControl, and investigated the effect of YB1 on miR-205/200b expression by qPCR analysis. As expected, regardless of the changes in Snail expression, YB1 knockdown remarkably increased the expression levels of pre- and mature miR-205 or miR-200b in MHCC97H, HCCLM3, HB611, and Huh7 cell lines compared to the control (Figure 5, Supplementary Figure S5). Taken together, these results suggest that YB1 could repress miR-205/200b expression partially in a Snail-independent manner in HCC cells.
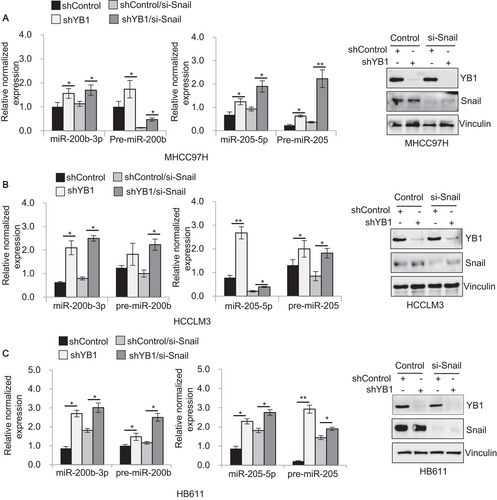
3.6 YB1 interacts with TUTs and influences miR-205/200b expression
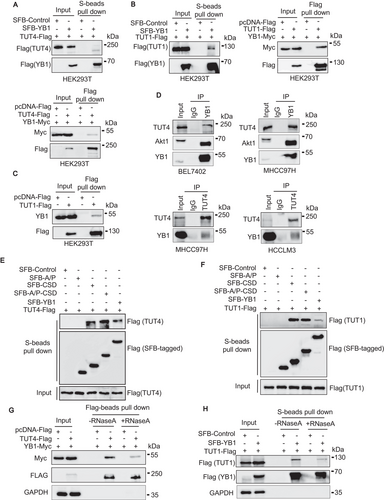
Some evidence has shown that TUTs influence miRNAs maturation via terminal uridylation of pre-miRNAs [41–43, 58, 59]. In this study, we found that YB1 could interact with TUT4 and TUT1 respectively by pull-down assays in HEK293T cells (Figure 6A–C). Furthermore, coimmunoprecipitation assays indicated that endogenous YB1 could interact with TUT4 in BEL7402, HCCLM3, and MHCC97H cell lines, respectively (Figure 6D). In addition, we found that all YB1 truncated constructs containing CSD exhibited strong associations with exogenous TUT4 and TUT1, suggesting that CSD may play crucial roles in YB1-TUT interactions (Figures 6E and 6F). To clarify whether RNAs contribute to the interactions of YB1 and TUT4/TUT1, we treated the cell lysates with RNase A and repeated pull-down experiments. We observed that RNase A treatment slightly weakened the interactions of YB1 and TUT4/TUT1 (Figures 6G and 6H). These results indicated that the interactions of YB1 and TUT4/TUT1 may be independent of RNAs. In addition, qPCR analysis demonstrated that either TUT4 or TUT1 overexpression remarkably enhanced the levels of precursor and mature miR-205/200b in HEK293T cells. However, cotransfection of YB1 with TUT4 or TUT1 reduced most of the increased miR-205/200b expression levels (Supplementary Figure S6). Collectively, these results indicate that YB1 could inhibit the activation of miR-205 and miR-200b by TUT4 or TUT1.
3.7 YB1 promotes cell invasion and tumor progression by regulating the miR-205/200b‒ZEB1 axis
To further confirm that YB1 promotes cell invasion by regulating the miR-205/200b‒ZEB1 axis, we silenced miR-205 and miR-200b with specific miRNA inhibitors in YB1 knockdown MHCC97H cells. As expected, YB1 knockdown significantly reduced the invasive cells compared with the shControl (Figure 7A). However, transfection of specific miR-205 or miR-200b inhibitors in MHCC97H cells stably expressing YB1 shRNA remarkably increased the invasive cells, suggesting that YB1 could induce cell invasion partially by downregulating miR-205 and miR-200b.
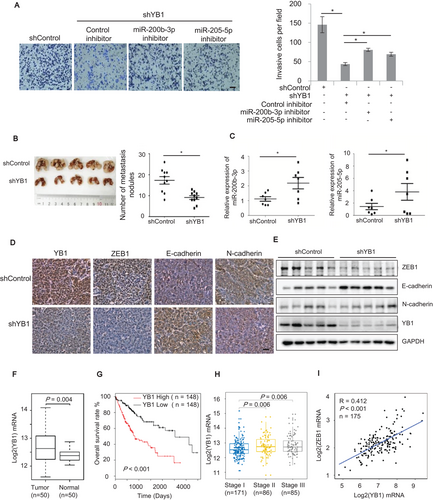
Next, we investigated whether YB1 could promote tumor metastasis in vivo. We inoculated MHCC97H cells stably expressing YB1 shRNA or shControl into BALB/C nude mice by tail vein injection. After 8 weeks, the nude mice were sacrificed and the lymph nodules derived from lung tissues were analyzed. As shown in Figure 7B, the nude mice injected with YB1 shRNA knockdown cells had fewer nodules than those injected with shControl cells, suggesting that YB1 could promote liver tumor metastasis in vivo. Notably, qPCR results demonstrated that the average expression levels of miR-205 and miR-200b were higher in the nodules derived from the YB1 shRNA knockdown group than those from the shControl group, suggesting that YB1 may suppress the expression of miR-205 and miR-200b in vivo (Figure 7C). In addition, the immunohistochemical analysis demonstrated that ZEB1 expression was lower in the nodules derived from the YB1 knockdown group than those from the shControl group (Figure 7D). The immunoblotting results further confirmed that YB1 was positively associated with ZEB1 expression and negatively correlated with E-cadherin expression (Figure 7E), which were consistent with the results in vitro (Figures 1A and 1B).
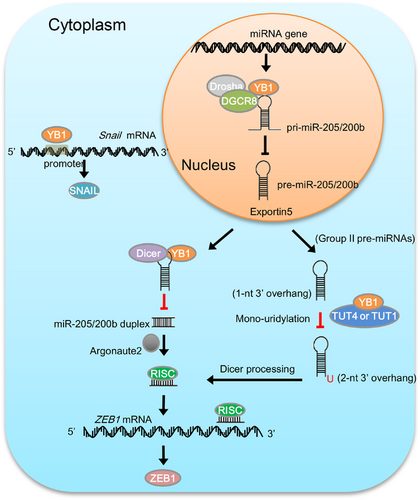
To investigate the relevance of our findings to human liver cancers, we detected the expression of YB1 and ZEB1 in HCC tissues. The results indicated that YB1 was significantly upregulated in cancer tissues compared with the adjacent non-cancer tissues, and its expression was positively associated with ZEB1 expression (Supplementary Figure S7A-B). Furthermore, we also analyzed gene expression data from The Cancer Genome Atlas (TCGA) [48, 49] and GTEx [51]. Consistent with our immunoblotting data (Supplementary Figure S7A-B), the expression of YB1 in the TCGA HCC cohort was significantly upregulated in the HCC tissues compared with the matched normal liver tissues (Figure 7F). Furthermore, the HCC patients with low levels of YB1 showed better overall survival than those with high YB1 expression levels (Figure 7G; 40% and 60% quantiles of YB1 expression values were used as the cutoffs for low and high expressions). In addition, the expression of YB1 in the early HCC aggressiveness stage was significantly lower than that in later stages (Figure 7H), and was positively correlated with ZEB1 expression in liver tissues (Figure 7I). Collectively, we postulate the working model by which YB1 promotes cell invasion and tumor metastasis through regulating miR-205/200b‒ZEB1 axis in HCC cells (Figure 8).
4 DISCUSSION
YB1 plays crucial roles in tumorigenesis and cancer progression through modulation of gene transcription and mRNAs translation [7, 10, 60]. However, whether YB1 contributes to malignant transformation via regulation of microRNA maturation remains largely unknown. In this study, we found that YB1 promoted cell migration and invasion by regulating miR-205/200b‒ZEB1 axis in HCC cells. Mechanistically, YB1 inhibited miR-205/200b maturation by interacting with microprocessors DGCR8 and Dicer, and subsequently activated ZEB1 expression and promoted cancer progression in HCC cells (Figure 8). This study highlights the implications that YB1 may play critical roles in human cancers by regulating miRNAs maturation and functions.
A previous study reported that YB1 induced EMT and enhanced metastatic ability by activation of the EMT inducer Snail in breast cancer [10]. Recently, another study revealed that Snail affected miRNAs expression in the early steps of metastasis in the colon cancer cell line HT29 [57]. To investigate whether YB1 regulates miR-205/200b expression dependent on Snail activation, we knocked down Snail in HCC cell lines stably expressing YB1 shRNA or shControl, and investigated the impact of YB1 on miR-205/200b expression by qPCR analysis. The results indicated that YB1 knockdown significantly enhanced miR-205/200b expression in MHCC97H, HCCLM3, HB611, and Huh7 cells regardless of the changes in Snail expression. These data suggest that YB1 may repress miR-205/200b expression partially in a Snail-independent manner. Collectively, this present study identified a previously undescribed mechanism through which YB1 could promote cell migration and invasion by regulating the miR-205/200b‒ZEB1 axis in HCC cells.
Dysregulation of miRNAs have been reported to contribute to tumorigenesis and progression in various cancers [15, 17, 18, 61]. The miR-205 and miR-200 family members are known to suppress cell EMT and tumor metastasis by inhibiting ZEB1 and SIP expression in cancer cells [16]. In this study, we found that YB1 inhibited miR-205 and miR-200b maturation through interacting with DGCR8 and Dicer. Notably, the miRNAs maturation is controlled by very complicated biogenesis machinery in cells. Microprocessor complexes Drosha and DGCR8 process primary miRNAs into precursor miRNAs in the nucleus, and ulteriorly Dicer engineered precursor miRNAs into mature miRNAs in the cytoplasm [23, 25–28]. Numerous studies have reported that RNA-binding proteins regulate specific miRNAs expression by interacting with the microprocessors or associated regulatory components, such as DDX1, Hippo, p68, and p72 [33-37]. Therefore, we assumed that interactions of YB1 with DGCR8 or Dicer could prevent them from recognizing and splicing primary transcripts or precursor miRNAs, thus leading to downregulation of mature miR-205/200b expression. To our knowledge, this is the first report that YB1 regulates miR-205/200b maturation and function by interacting with the microprocessors DGCR8 and Dicer.
Some RNA-binding proteins have been reported to modulate miRNA processing efficiency in a sequence or structure-dependent manner [62]. In this study, we identified that YB1 bound to the terminal loop of pre-miR-205 and pre-miR-200b. A previous study has reported that YB1 regulated miR-29b-2 biogenesis by recognizing a specific RNA motif of “UYAUC” at the loop region [5]. Notably, the loop sequences of pre-miR-205 and pre-miR-200b are “AUACC” and “AGUCA” respectively, which contain the variable forms of “AUC” (UAC and UCA) compared to pre-miR-29b. Both pull-down and RNA immunoprecipitation assays identified that YB1 could bind more efficiently to the wild-type pre-miR-205 and pre-miR-200b than to their mutant constructs. These results suggest that the loop sequence and structure may affect the special interaction between YB1 and pre-miRNAs. It is to be further investigated whether other pre-miRNAs with the variable RNA motif (AUC, UAC, and UCA) at the terminal loop region could be regulated by YB1 via the same mechanism.
In this study, we also identified that YB1 could physically interact with TUT4 and TUT1. TUT4 has been reported to interact with Lin28 and suppress let-7 miRNA biogenesis by terminal uridylation [41, 42]. We hypothesized that YB1 could repress miR-205/200b maturation in combination with TUTs via the same mechanism. However, qPCR analysis indicated that either TUT4 or TUT1 overexpression remarkably enhanced the expression of miR-205 and miR-200b, whereas the elevated miR-205/200b transcripts were largely reduced by cotransfecting YB1 with TUT4 or TUT1. The results indicated that YB1 could inhibit the activation of miR-205/200b expression by TUT4 and TUT1. Notably, terminal uridylyl transferases TUT4, TUT7, and TUT2 (GLD2/PAPD4) have been reported to mediate mon-uridylation of pre-miRNAs and facilitate Dicer processing of group II let-7 miRNAs biogenesis [58, 59]. The structures of human pre-miR-205 and pre-miR-200b have 1-nt at 3′ overhang, which are supposed to belong to group II miRNAs [43, 58]. TUT4 overexpression may contribute to Dicer processing of pre-miR-205 and pre-miR-200b via terminal mon-uridylation. However, YB1 may block TUT4 binding to pre-miRNAs via competing for the binding sites, thus leading to downregulation of mature miR-205 and miR-200b. Besides, previous studies have reported that TUT1 contributes to U6 small nuclear RNA splicing via oligo-uridylylation [45, 63]. In this study, we found that TUT1 could enhance miR-205/200b expression in cancer cells. However, the detailed molecular mechanisms of YB1 how regulates TUTs and their functions on miRNAs biogenesis in human cancers should be investigated more in the future.
5 CONCLUSIONS
Our study unveils an undescribed mechanism through which YB1 promotes HCC cell migration and tumor metastasis by regulating miR-205/200b‒ZEB1 axis in a Snail-independent manner. Here, we identified that YB1 enhanced ZEB1 expression by inhibiting miR-205/200b maturation through interacting with the microprocessors DGCR8 and Dicer as well as TUT4 and TUT1. Statistical analyses on the gene expression data from GTEx liver tissues and TCGA HCC cohort further confirmed that the expressions of YB1 and ZEB1 were positively associated and a high YB1 expression corresponded to poor clinical prognosis, suggesting that YB1 could serve as a potential therapeutic target in liver cancers. Collectively, this study uncovers a novel function of YB1 as a specific regulator of miRNA biogenesis, which highlights that YB1 may play critical functions in cancer biology via miRNAs-mediated gene regulation.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Clinical Experiment Ethics Committee of the First Affiliated Hospital of Dalian Medical University. All patients had signed informed consents and agreed to donate their tissues for the research. The independent fresh HCC and adjacent non-tumor tissues were obtained from the patients during the surgical resection. The animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of Dalian Medical University.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
We thank all members of Dr. Hai-long'Piao's laboratory for helpful discussion. We thank V Narry Kim (School of the Biological Sciences, Seoul National University, Seoul, Korea), Sandra E Dunn (Phoenix Molecular Designs, Vancouver, British Columbia, Canada), and Kimitoshi Kohno (Asahi-Matsumoto Hospital, Kitakyushu, Japan) for kindly providing the plasmids used in this study. This study was supported by National Natural Science Foundation of China grants (81672440, 31701156, 81972625), Innovation program of science and research from the DICP (DICP ZZBS201803), Department of Liaoning Science and Technology, titled with The Construction of Liaoning Cancer Research Center (Lung Cancer) (1564992449013) and titled with Precise diagnosis and treatment, and optimization of a clinical pathway for malignant tumor based on molecular markers-the research of precise treatment and optimization of a clinical pathway for lung cancer (2019020176-JH1/103-02), Central financial fund for promoting medical service and safeguarding capability (Capability construction of medical and health organizations)-a subsidy to the Construction of Provincial Key Specialty, and Research grant to introduced talents of Liaoning Cancer Hospital.
AUTHORS’ CONTRIBUTIONS
X.L. and H.P. conceived the project. H.P. supervised the project. X.L. designed and performed most of the experiments. X.L., D.C. and H.P. analyzed data. D.C. performed computational data analysis. H.C., W.W., Y.L., Y.W., C.D., Z.N., X.G., W.O., J.L., H.Q., X.L., A.L., and T.X. provided significant intellectual input. X.L., H.L., D.C., T.X. and H.P. wrote the manuscript with input from all other authors.




