Click-iG: Simultaneous Enrichment and Profiling of Intact N-linked, O-GalNAc, and O-GlcNAcylated Glycopeptides
Graphical Abstract
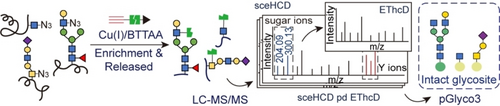
A glycoproteomics platform, Click-iG, is reported that integrates metabolic labeling of glycans with clickable unnatural sugars, an optimized MS method, and a tailored version of pGlyco3 software to enable simultaneous enrichment and profiling of three types of intact glycopeptides: N-linked, mucin-type O-linked, and O-GlcNAcylated glycopeptides.
Abstract
Proteins are ubiquitously modified with glycans of varied chemical structures through distinct glycosidic linkages, making the landscape of protein glycosylation challenging to map. Profiling of intact glycopeptides with mass spectrometry (MS) has recently emerged as a powerful tool for revealing matched information of the glycosylation sites and attached glycans (i.e., intact glycosites), but is largely limited to individual glycosylation types. Herein, we describe Click-iG, which integrates metabolic labeling of glycans with clickable unnatural sugars, an optimized MS method, and a tailored version of pGlyco3 software to enable simultaneous enrichment and profiling of three types of intact glycopeptides: N-linked, mucin-type O-linked, and O-GlcNAcylated glycopeptides. We demonstrate the utility of Click-iG by the identification of thousands of intact glycosites in cell lines and living mice. From the mouse lung, heart, and spleen, a total of 2053 intact N-glycosites, 262 intact O-GalNAc glycosites, and 1947 O-GlcNAcylation sites were identified. Click-iG-enabled comprehensive coverage of the protein glycosylation landscape lays the foundation for interrogating crosstalk between different glycosylation pathways.
Conflict of interest
A patent application covering the use of Click-iG has been filed, in which Peking University is the applicant and X. C. and J. L. are the inventors.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.