Local Transient Unfolding of Native State PAI-1 Associated with Serpin Metastability†
The authors thank Prof. Elizabeth Komives for comments to the manuscript. M.B.T., J.B.M., T.J.D.J., and P.A.A. were supported by a grant from the Danish Research Council for Technical Sciences and Production (grant number 09-072885). M.B.T. was additionally supported by grants from The Lundbeck Foundation (grant number R18-A11217) and The Carlsberg Foundation (grant number 2012_01_0369).
Graphical Abstract
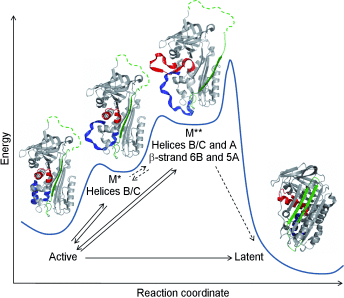
Serpin proteins are prone to pathological conformational change, for instance by conversion into an inactive, so-called latent form. By using advanced hydrogen/deuterium-exchange mass spectrometry, transient unfolding of a serpin is shown under native conditions. Based on these observations, a new mechanism (see picture) is proposed.
Abstract
The metastability of the native fold makes serpin (serine protease inhibitor) proteins prone to pathological conformational change, often by insertion of an extra β-strand into the central β-sheet A. How this insertion is made possible is a hitherto unresolved question. By the use of advanced hydrogen/deuterium-exchange mass spectrometry (HDX-MS) it is shown that the serpin plasminogen activator inhibitor 1 (PAI-1) transiently unfolds under native condition, on a second-to-minute time scale. The unfolding regions comprise β-strand 5A as well as the underlying hydrophobic core, including β-strand 6B and parts of helices A, B, and C. Based thereon, a mechanism is proposed by which PAI-1 makes transitions through progressively more unfolded states along the reaction coordinate to the inactive, so-called latent form. Our results highlight the profound utility of HDX-MS in detecting sparsely populated, transiently unfolded protein states.