A Strategy for Functional Proteomic Analysis of Glycosidase Activity from Cell Lysates†
The authors thank S. G. Withers for samples of recombinant Agrobacterium sp. β-glucosidase and Xanthomonas manihotis β-galactosidase, and H. C. Hang for useful discussions. D.J.V. thanks the Canadian Institutes of Health Research for a fellowship. The authors thank A. Debowski for technical assistance. This research was supported by a grant to C.R.B. from the National Institutes of Health (Grant no. GM066047), a President's Research Grant from Simon Fraser University, and a grant to D.J.V from the Natural Sciences and Engineering Research Council of Canada. The center for New Directions in Organic Synthesis is funded by Bristol–Myers Squibb as a supporting member and Novartis as a sponsoring member.
Graphical Abstract
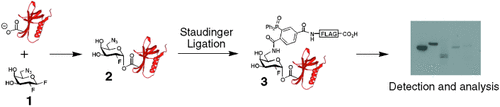
A multipurpose flag: The inactivator 1 covalently labels the catalytic nucleophiles of retaining β-glycosidases to form species 2. The small azide group allows labeling of enzymes with sterically congested active sites. Staudinger ligation of 2 with phosphine–FLAG yields adduct 3, which can be used to detect and profile retaining β-glycosidase activities in complex mixtures.