Congenital Titinopathy: Comprehensive Characterization of the Most Severe End of the Disease Spectrum
Titin Research Consortium collaborators are listed in Appendix.
Abstract
Congenital titinopathy has recently emerged as one of the most common congenital muscle disorders.
Objective
To better understand the presentation and clinical needs of the under-characterized extreme end of the congenital titinopathy severity spectrum.
Methods
We comprehensively analyzed the clinical, imaging, pathology, autopsy, and genetic findings in 15 severely affected individuals from 11 families.
Results
Prenatal features included hypokinesia or akinesia and growth restriction. Six pregnancies were terminated. Nine infants were born at or near term with severe-to-profound weakness and required resuscitation. Seven died following withdrawal of life support. Two surviving children require ongoing respiratory support. Most cohort members had at least 1 disease-causing variant predicted to result in some near-normal-length titin expression. The exceptions, from 2 unrelated families, had homozygous truncating variants predicted to induce complete nonsense mediated decay. However, subsequent analyses suggested that the causative variant in each family had an additional previously unrecognized impact on splicing likely to result in some near-normal-length titin expression. This impact was confirmed by minigene assay for 1 variant.
Interpretation
This study confirms the clinical variability of congenital titinopathy. Severely affected individuals succumb prenatally/during infancy, whereas others survive into adulthood. It is likely that this variability is because of differences in the amount and/or length of expressed titin. If confirmed, analysis of titin expression could facilitate clinical prediction and increasing expression might be an effective treatment strategy. Our findings also further-support the hypothesis that some near-normal-length titin expression is essential to early prenatal survival. Sometimes expression of normal/near-normal-length titin is due to disease-causing variants having an additional impact on splicing. ANN NEUROL 2025;97:611–628
The TTN gene encodes titin—the largest protein in nature.1 In skeletal and cardiac muscle, 2 titin molecules pair to span the full length of the sarcomere forming a continuous elastic myofilament that provides a scaffold for sarcomere assembly during muscle development,2-4 modulates sarcomeric tension during contraction, and serves as an important mechanosensing and signaling hub (reviewed in Gautel et al5).
As a result of the increasing use of massively parallel sequencing (MPS) in diagnostics, it has become apparent that disease-causing TTN variants (sometimes referred to as pathogenic/deleterious variants or mutations) are responsible for several important skeletal muscle and cardiac disorders.
Heterozygous (de novo or dominantly inherited) nonsense, frameshift, and canonical splice site-altering TTN variants are the most common cause of familial/genetic adult-onset dilated cardiomyopathy (DCM).6-8 In addition, heterozygous disease-causing variants in 2 specific regions of TTN are responsible for 2 dominantly inherited skeletal muscle titinopathies: (1) tibial muscular dystrophy (TMD: final exon variants)9, 10 and (2) hereditary myopathy with early respiratory failure (HMERF: exon 344 missense variants).11, 12
Multiple different recessive skeletal muscle titinopathies have also been described, under different clinical labels.13-20 These conditions are increasingly considered clinical variations of the same disorder; recessive titinopathy (RT). With the increasing use of MPS in diagnostics, RT has emerged as a common cause of prenatal-, childhood-, and adolescent-onset skeletal muscle disease, frequently complicated by cardiac complications. For example, RT is now the second most common genetic diagnosis in Australian PathWest gene panel-analyzed myopathy cases (M.D., panel lead, reporting limited to cases with truncating and/or convincingly pathogenic splice-altering variants).
Congenital titinopathy, the most severe form of RT, manifests in utero or during infancy16-20 and can result in prenatal or early infant death. However, severity is highly variable and well over 20 reported congenital titinopathy cases have survived into adulthood (>18 years; oldest published congenital/infant-onset case is 57 years).16, 20-25 The biological mechanisms that underpin these marked differences in severity remain unclear.
Our understanding of the RT clinical spectrum has expanded rapidly over the past decade. However, our knowledge of titin transcript and protein isoform biology remains limited. In humans, there is currently only 1 characterized (canonical) titin mature skeletal muscle isoform: N2A.1, 26 However, multiple streams of evidence suggest that additional yet-to-be-characterized human fetal and postnatal (mature) muscle isoforms exist.28-30 In mature cardiac muscle, N2B and N2BA are 2 longest and most abundant titin isoforms. There are also several smaller cardiac isoforms (Novex 1, 2, 3).27
It increasingly appears that analysis of the isoform-level impact(s) of TTN disease-causing variants may facilitate prediction of clinical outcomes. For instance, Roberts et al7 showed that heterozygous truncating and canonical splice site TTN disease-causing variants that impact both N2B and N2BA are more likely to result in adult-onset DCM than other TTN disease-causing variants. Congenital titinopathy cases with biallelic TTN disease-causing variants that both impact N2BA and N2B also appear more likely to develop cardiac complications.20
Our study aims to comprehensively characterize the clinical and genetic findings in congenital titinopathy cases at the most extreme end of the severity spectrum and consider explanations for the severity of these cases.
Methods
The project was approved by the Sydney Children's Hospitals Network Human Research Ethics Committee (2019/ETH11736) and by other researchers' review boards. Consent for research participation and use of photographs was obtained from parents/legal guardians.
Only severely affected cases with convincingly pathogenic truncating and/or splice-altering TTN disease-causing variants confirmed to be in trans by parental segregation studies were included in this study. “Severely affected” was defined as having pronounced in utero features (absent/minimal fetal movements, multiple contractures and/or hydrops) and/or severe-to-profound hypotonia and respiratory insufficiency at birth necessitating immediate resuscitation and prolonged intensive respiratory support (>4 weeks or until death). Cases with difficult-to-interpret missense variants or incomplete segregation results were excluded.
The study included published and unpublished data from Families AUS1, UK1, and BEL1, reported previously as Family 6/AUS005, Family 17/UK0001, and Family 18/BEL0001.20
All disease-causing variants were identified via panel, whole exome or whole genome sequencing and reported according to Human Genome Variation Society recommendations referencing the inferred complete TTN metatranscript (ENST00000589042; NM_001267550.1; LRG391_t1). Exons were numbered 1–364 according to the Locus Reference Genomic (LRG) schema.32
The Leiden Muscular Dystrophy pages and Clinvar were interrogated to identify previously reported disease-causing variants. The frequency of each disease-causing variant was determined using the Genome Aggregation Database (gnomAD).33, 34 The protein-level impact(s) of each disease-causing variant was predicted using Alamut Visual. Cardiodb was consulted to determine if disease-causing variant-impacted exons were included in N2A (skeletal), N2B (cardiac), and/or N2BA (cardiac) isoform transcripts.
The predicted degree of nonsense mediated mRNA decay (NMD) triggered by each truncating disease-causing variant was determined using pre-computed NMDetective-A scores31 according to the position of the termination codon (sequential exon usage assumed for termination codons not in same exon as frameshift disease-causing variants).
Percent spliced in index (PSI)35 (exon usage) values for disease-causing variant-impacted exons were calculated using previously published methods.7 Fetal and pediatric skeletal muscle PSI values were generated from ribosomal RNA depleted 150 base pair single end Illumina RNA-seq data from 5 control fetal/neonatal samples and 14 control pediatric samples. Adult skeletal muscle (gastrocnemius) PSI values were calculated from poly-A selected-76 bp paired end Illumina GTEx36 RNA-seq data from 196 individuals without features of a neuromuscular disorder or a history of treatment or lifestyle factors that might impact muscle health. Cardiac muscle PSI values were previously published data, which were generated from 84 end-stage dilated cardiomyopathy patient samples.7
ESEFinder37 was used to identify disease-causing variants that altered exonic splicing enhancer (ESE) motifs and were, therefore, predicted to have additional splice impact(s). The strength and relative positioning of surrounding splice signals were analyzed to determine the likely splice impact(s) of ESE-altering disease-causing variants.
The minigene assay used to confirm the additional predicted splice impacts of the Family SRI1 homozygous nonsense disease-causing variant was performed as previously described38, 39 with slight modifications (Data S2). Sashimi plots were generated using ggsashimi.40
Cardiac and disease-causing variant data from members of this cohort and an additional 50 published families (Table S3) were stratified according to the cardiac status of affected members (“Yes” if congenital and/or non-congenital cardiac abnormalities reported in at least 1 RT case from that family), and whether each had (1) two N2B/N2BA-impacting disease-causing variants or (2) other combinations of disease-causing variants.
Results
Families
This international multicenter study comprises 15 severely affected fetuses and infants from 11 unrelated families. Antenatal information was available for 14 of 15 cases. Only autopsy data was available for MAL1.II.4.
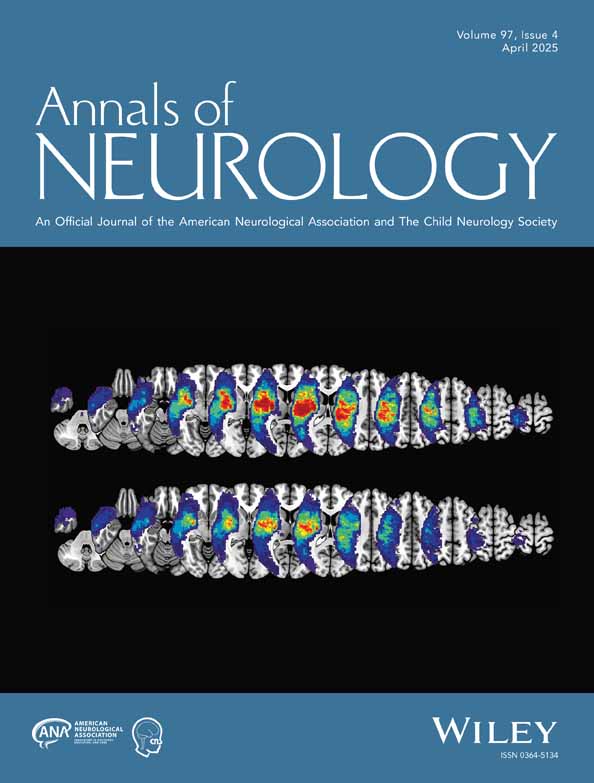
Family ethnicity was diverse. Family pedigrees are shown in Fig 1A.
Six families (MAL1, UK2, NL1, NL2, SRI1, and UK4) had a history of more than 1 severely affected child, with a consistently severe presentation within each sibship.
Eight families included ≥1 affected child who survived until birth, confirming that the combination of disease-causing variants present was compatible with in utero survival.
Overview of Cases
Six pregnancies had been terminated (11 weeks to 27 + 5 weeks gestation) because of severe fetal hypokinesia or akinesia, limb contractures, and/or hydrops fetalis. The remaining 9 infants were born alive at or after 35 weeks gestation. Seven of these infants subsequently died from cardiorespiratory compromise between days 1 and 44 of life. Two survived beyond infancy.
Features: Terminated Pregnancies
Table 1 describes the in utero imaging and autopsy features present in the 6 cases who did not survive to term. Six pregnancies in 4 families were terminated because of the presence of severe abnormalities and poor prognosis. Severe fetal hypokinesia or akinesia was present in all cases and detected as early as 11 weeks gestation. Additional abnormal in utero features included limb contractures and/or talipes (4/6, involving all limbs in all 4), hydrops fetalis (3/6 cases), intrauterine growth retardation (2/6), and increased nuchal translucency (2/6). No cardiac abnormalities were detected antenatally.
| Patient information | NL1.II.2 | NL1.II.3 | NL2.II.1 | NL2.II.3 | UK3.II.1 | UK4.II.5 | Total |
|---|---|---|---|---|---|---|---|
| Gender of fetus | Male | Male | Female | Male | Female | ||
| Gestation at TOP (weeks + days) | 11 | 20 | 14 | 11 | 27+5 | 22 | |
| In utero history | |||||||
| Decreased fetal movements | + | + | + | + | + | + | 6/6 |
| Hydrops fetalis | + | + | + | − | − | − | 3/6 |
| Increased nuchal translucency | + | + | − | − | 2/4 | ||
| Contractures/talipes on US | + | + | − | − | + | + | 4/6 |
| Intrauterine growth retardation | + | + | − | − | − | − | 2/6 |
| Polyhydramnios | − | − | − | − | + | − | 1/6 |
| Autopsy features | |||||||
| Arthrogryposis multiplex congenita | + | + | + | + | + | 5/5 | |
| Hydrops fetalis | + | + | + | − | − | 3/5 | |
| Fetal cystic hygroma | + | + | + | − | − | 3/5 | |
| Thoracic hypoplasia | + | + | − | + | − | 3/5 | |
| Muscle hypoplasia | − | − | − | + | + | 2/5 | |
| Microretrognathia | + | + | + | + | + | 5/5 | |
| Low-set ears | + | + | + | − | + | 4/5 | |
| Abnormal facial shape | + | + | − | − | − | 2/5 | |
| Decreased palmar creases/abnormal finger flexion creases | + | − | − | + | 2/4 | ||
| Additional hand/foot abnormalities | Syndactyly | − | − | Congenital finger flexion contractures, rocker bottom foot, sandal gap, toe oligodactyly | Hand clenching | 3/5 | |
| Additional features | Gastroschisis | Cleft palate | Anal atresia | Hypertelorism | 4/4 | ||
- In the last column (Total), the denominator is the number of cohort members with data provided for that feature. If no data was available regarding a specific feature the entry has been left blank.
- TOP = termination of pregnancy, US = ultrasound.
For all 6, there were documented in utero findings, and 5 underwent autopsy. All 6 had features consistent with fetal akinesia deformation sequence41 including contractures involving all 4 limbs, facial dysmorphisms (eg, micro/retrognathia [5/5 with autopsy findings], low set ears [4/5]), and a narrow/hypoplastic thorax (3/5). A cystic hygroma, significant muscle hypoplasia, and decreased palmar and/or finger creases consistent with a paucity of in utero hand/finger movements were noted in a subset of cases (3/5, 2/5, and 2/5, respectively). Cleft palate, a feature noted in previously reported cases,18, 20 was present in 1 fetus. Gastroschisis and anal atresia were each present in 1 infant.
Features: Infants Born At or Near Term
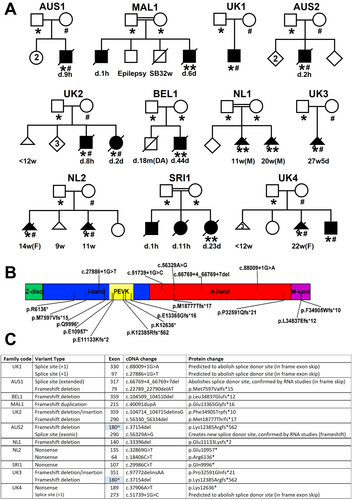
Table 2 describes the clinical features present in the 9 infants who survived until birth and the autopsy features in 5 of these infants, with more detailed summaries provided in Tables S4 (clinical features) and S5 (autopsy features). Figure 2 shows images of 3 severely affected infants who died shortly after birth.
| Patient | UK1.II.1 | AUS1.II.3 | BEL1.II.2 | MAL1.II.4 | UK2.II.5 | UK2.II.6 | SRI1.II.3 | AUS2.II.3 | UK4.II.6 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Male | Male | Male | Female | Female | Male | Male | |
| In utero history | ||||||||||
| Hypokinesia or akinesia | + | + | + | + | + | + | + | + | 8/8 | |
| Contractures/talipes | − | + | − | − | − | − | + | + | 3/8 | |
| IUGR | − | − | + | − | − | + | − | − | 2/8 | |
| Polyhydramnios | − | − | − | + | + | + | + | − | 4/8 | |
| Delivery history | ||||||||||
| Gestational age | Term | 38 | 39+5 | 39 | 38+5 | 35+1 | 37 | 35 | 37+2 | |
| Birth weight (kg) | 2.165 | 3.212 | 2.230 | 2.320 | 2.315 | |||||
| Breech presentation | − | + | − | + | + | − | + | − | − | 4/9 |
| Caesarean section | + | + | − | + | + | + | + | − | − | 6/9 |
| Respiratory distress | + | + | + | + | + | + | + | + | 8/8 | |
| Neonatal asphyxia | + | − | − | − | + | − | − | + | 3/8 | |
| Muscular and osteoarticular features | ||||||||||
| Neonatal hypotonia and weakness | + | + | + | + | + | + | + | + | + | 9/9 |
| Muscular hypoplasia | − | − | + | − | − | − | + | + | 3/8 | |
| Arthrogryposis | − | + | + | + | + | + | + | + | + | 8/9 |
| Fractures long bones | − | + | − | + | − | − | − | + | + | 4/9 |
| Cardiac features | ||||||||||
| Atrial septal defect | − | − | + | − | + | + | − | 3/7 | ||
| Ventricular septal defect | + | − | − | − | − | − | − | 1/7 | ||
| Patent ductus arteriosus | − | − | − | − | + | + | − | 2/7 | ||
| Dysmorphic features | ||||||||||
| High, narrow palate | − | + | + | + | + | + | − | 5/7 | ||
| Myopathic facies | + | − | + | 2/3 | ||||||
| Low-set ears | − | − | + | + | + | + | − | 4/7 | ||
| Narrow mouth | − | + | − | − | − | − | + | − | 2/8 | |
| Micrognathia or retrognathia | − | − | − | + | + | − | + | + | + | 5/9 |
| Abnormality of the neck | − | − | − | + | + | + | − | 3/7 | ||
| Hydrops fetalis | − | − | − | − | − | + | − | 1/7 | ||
| Cryptorchidism | − | + | − | + | + | + | 4/6 | |||
| Autopsy features | ||||||||||
| Pulmonary hypoplasia | + | + | ± | − | 2/4 | |||||
| Pleural effusion | + | − | + | + | 3/4 | |||||
- In the last column (total), the denominator is the number of cohort members with data provided for that feature. If no data was available regarding a specific feature the entry has been left blank. More detailed information regarding clinical and autopsy findings is provided in Tables S4 and S5, respectively. Note 1: Gestational age is reported in weeks + days. Note 2: pulmonary hypoplasia is defined as lung weight/body weight ratio <1.2%.
- IUGR = intrauterine growth retardation; NA = not assessed; US = ultrasound.
Pregnancy data were available for 8 of the 9 infants who were born alive at or near term (only autopsy data for 9th). All 8 had reduced in utero movements noted at or after 20 weeks gestation. A subset (3/8) had additional limb contractures and/or talipes (involving all 4 limbs in 2/3), intrauterine growth retardation (2/8), and/or increased nuchal translucency (1/8) on prenatal imaging. Polyhydramnios developed at or after 28 weeks gestation in 4 of 8 cases. None had hydrops fetalis.
Six infants were delivered by Caesarean section because of breech presentation (4/6) and/or fetal distress. All had either no respiratory movements or developed marked respiratory distress following delivery. All required immediate resuscitation and mechanical ventilation. Table S4 shows additional birth history information.
Three infants (UK1.II.1, BEL1.II.2, and UK4.II.6) had brain magnetic resonance imaging (MRI) features consistent with hypoxic brain injury following traumatic deliveries. UK1.II.1 subsequently developed seizures and was treated with therapeutic hypothermia and anticonvulsants. A fourth infant (UK2.II.5) had signs of severe hypoxic–ischemic encephalopathy and died at 8 hours of age from severe respiratory insufficiency.
All infants born alive had severe-to-profound axial and limb hypotonia, absent proximal antigravity movements, reduced or absent distal antigravity movements, and congenital contractures involving all 4 limbs (Fig 2). Four had bilateral talipes equinovarus (Fig 2). Three had decreased or absent palmar and/or phalangeal creases.
A high-arched palate was a frequent finding (5/6). Ophthalmoplegia was consistently absent. Mild facial weakness or myopathic facies were present in 2 cases. Micro/retrognathia, low set ears, and a narrow mouth were each present in more than 1 case (5, 3, and 2 cases, respectively). Four cases had neck abnormalities (webbing, cystic swelling, increased skin, and short neck). All infants who survived beyond the first few days of life had significant feeding difficulties.
Four infants (BEL1.II.2, UK1.II.1, UK2.II.6, and AUS2.II.3) had atrial and/or ventricular septal defects. Two (BEL1.II.2, UK2.II.6) had additional pulmonary hypertension. In UK2.II.6, this resulted in right-sided cardiac failure. In BEL1.II.2, echocardiogram-confirmed pulmonary hypertension resolved within 1 month.
Four infants (AUS1.II.3, AUS2.II.3, UK2.II.5, and UK2.II.6) died from profound respiratory insufficiency during the first 48 hours of life. The 5 infants who survived beyond 48 hours (UK1.II.1, BEL1.II.2, MAL1.II.4, SRI1.II.3, and UK4.II.6) remained dependent on mechanical ventilation (weeks-months). Three subsequently died following withdrawal of respiratory support because of absence of clinical improvement or deterioration.
UK1.II.1 (age, 3 years) remains alive following a period of intensive medical intervention throughout early childhood. He remains dependent on tracheostomy-administered ventilation and is hospitalized several times a year for lower respiratory tract infections. He is severely hypotonic, weak (gross motor function classification system level 5), and is fed via gastrostomy tube. He is unable to vocalize, but is socially interactive.
UK4.II.6 (age, 16 months) required mechanical ventilation for 109 days, remains dependent on nocturnal bi-level positive airway pressure ventilation, and is awaiting insertion of a gastrostomy tube. He has antigravity finger and wrist movements (contractures restrict finger movements) and can reach and grasp, but has subgravity hip and knee strength. Lower limb movement is limited to minimal kicking. His congenital fractures have healed. However, he is osteopenic, with thin bones and is being monitored closely for bone health-related complications.
The 2 infants (BEL1.II.2, UK1.II.1) with available creatine kinase (CK) levels had normal results. Three infants underwent electromyography (EMG), nerve conduction studies (NCS), and/or repetitive nerve stimulation testing (RNS) with variable results (Table S4).
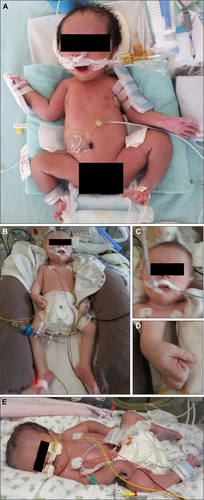
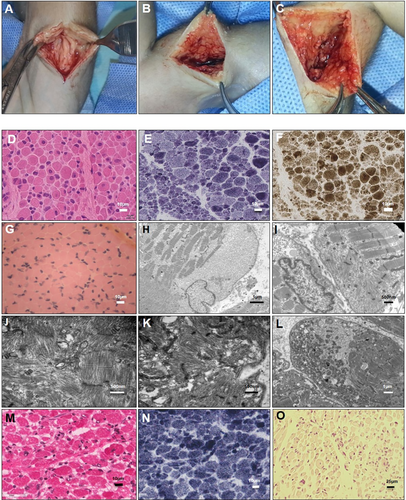
All 5 infants who were live-born and later underwent autopsy had dysmorphic and/or myopathic facial features and arthrogryposis multiplex congenita. One infant (MAL1.II.4) had complete absence of multiple muscle groups including sternocleidomastoid, right biceps, and thigh muscles (Fig 3A–C). Intercostal muscles were markedly atrophic. The diaphragm was macroscopically normal. There was also fibrous tissue replacement of the cervical spinal cord of uncertain significance.
Pulmonary hypoplasia (lung weight/body weight <1.2%42) was present in 2 infants. All 4 male infants had undescended testes.
Imaging Abnormalities
Figure 4 shows prenatal and postnatal MRI, ultrasound, and/or X-ray imaging studies of 4 severely affected infants; in utero ultrasound and MRI images for AUS2.II.3; and postnatal X-ray and/or MRI images for AUS1.II.3, MAL1.II.4 and UK4.II.6. Table 1 (fetal cases) and Table 2 (surviving infants) summarizes the imaging abnormalities noted in each of these cases.
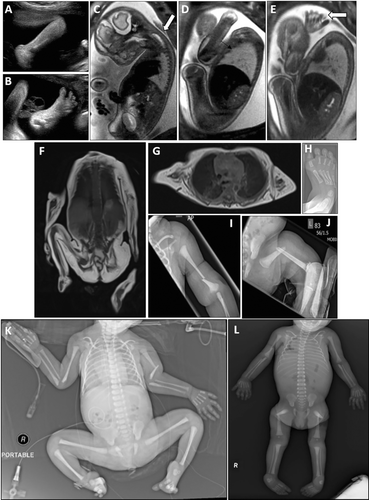
These images show evidence of markedly reduced in utero movements, limb and toe flexion deformities, and limb girdle, hand, paravertebral, neck, tongue, and/or facial muscle atrophy. Premortem and postmortem X-ray findings included diaphyseal and metaphysical fractures of limb bones (radius, humerus, femur, and distal tibial), which were occasionally bilateral (eg, MAL1.II.4: both humeri in [Fig 3K]). Some fractures looked recent, with no evidence of repair, whereas others (eg, the distal radius and ulna fractures in AUS1.II.3) (Fig 3L), appeared more longstanding.
Muscle Histopathology
Premortem skeletal muscle histopathology results were available for 5 live-born infants. Postmortem skeletal muscle histopathology results were available from 1 fetus terminated at 27 + 5 weeks gestation (eg, Fig 3, summary of findings, Table 3).
| Patient | Age at biopsy | Site of biopsy | Light microcopy findings | Ultrastructural (EM) findings |
|---|---|---|---|---|
| UK1.II.1 | 2 months 13 days (born at term) | Quad |
|
Not undertaken |
| AUS1.II.3 | 38/40 (before death at 8 hours of age) | Quad |
|
|
| BEL1.II.2 | Day 7 (born at 39+5/40) | Quad |
|
|
| UK2.II.6 | Not known | Not known |
|
|
| AUS2.II.3 | Day 1 (born at 35/40) | Gastrocnemius |
|
|
| UK3.II.1 | 27+5/40 (TOP) | Quad, triceps, psoas |
|
Not undertaken |
- EM = electron microscopy; FSV = fiber size variation; IN = internalized nuclei; Quad = quadriceps; TOP = termination of pregnancy.
Histopathological changes were variable both in severity and nature ranging from a dystrophic appearance (AUS 1.II.3) to myopathic and milder nonspecific abnormalities. All biopsies showed 1 or more of the following pathological features: atrophy and/or fiber-size variation (all 6/6 biopsies), increased internalized nuclei (3/6), core-like structures (1/6), and cap-like lesions (2/6).
Features suggestive of ongoing degeneration and/or regeneration were noted in several biopsies. Ultrastructural examination revealed focal loss or disorganization of myofibrillar structure in 4 biopsies, including 1 with only minimal light microscopy changes. Striking central and circumferential peripheral mitochondrial accumulations were visible on oxidative enzyme-stained sections from 2 biopsies (from AUS1.II.3, UK2.II.6).
Cardiac Findings in Carrier Relatives
Cardiac screening of carrier relatives was incomplete (undertaken in 4/22 parents and no carrier siblings). Furthermore, many carrier family members remained under the age of 40 and may, therefore, be yet to manifest TTN-related cardiac pathology (risk increases with age). In the context of these limitations, there was no history of DCM among first-degree relatives of cohort members. The mother of AUS2.II.3 had been diagnosed with atrial fibrillation before the age of 40.
Disease-Causing Variants
The location of the 18 cohort member disease-causing variants (Fig 1C) mapped against a schematic representation of the domains encoded by the complete inferred TTN metatranscript (Refseq transcript NM_001267550.1) is shown in Fig 1B. A comprehensive description of each disease-causing variant is provided in Table S1A, along with patient and parental cardiac findings.
Ten disease-causing variants were novel. All disease-causing variants were absent from gnomAD with the single exception of one very low frequency disease-causing variant (AUS1: p.Met7597Valfs*15: frequency: 1/244158). Four disease-causing variants were nonsense, 9 were frameshift, and 3 were canonical splice site disease-causing variants predicted to cause in-frame skipping of a single exon. Two were non-canonical splice site disease-causing variants. The first (in AUS2) altered the final base of an exon. Muscle complementary DNA (cDNA) studies confirmed that this disease-causing variant resulted in the creation of a new donor splice site and subsequent frameshift (data not shown). The second (in AUS1) was an extended splice site deletion shown by cDNA studies to result in in-frame skipping of exon 317 as previously described.20
Most disease-causing variants were in N2A exons with PSI values of 100% (Table S1A). Three disease-causing variants from 4 cohort families (MAL1, AUS2, UK3, and UK4) were within non-N2A I-band exons. This included an exon 180 p.Lys12385Argfs*562 disease-causing variant identified in 2 unrelated families (AUS2, UK3). All 3 exons that contained non-N2A disease-causing variants had fetal or neonatal PSI values that were similar to or greater than pediatric and adult PSI values (Table S1A), suggesting that these exons are included within a significant subset of fetal isoform transcripts.
Previously Unrecognized Splicing Impact(s)
Seven nonsense and frameshift disease-causing variants were predicted to have an additional previously unrecognized impact on splicing (Table S1A: columns V and W).
The Family SRI1 homozygous nonsense disease-causing variant in exon 107 (PSI 100% in fetal or neonatal, pediatric, and adult muscle) was predicted to result in the loss of a strong serine and arginine rich splicing factor 5 (SRSF5) splicing motif (Fig 5A). Minigene splicing analysis confirmed that this disease-causing variant results in markedly increased usage of a cryptic splice acceptor site within exon 107 (Fig 5C) and in-frame loss of 261 base pairs of exon 107, including the premature termination codon introduced by the disease-causing variant (Fig 5B).
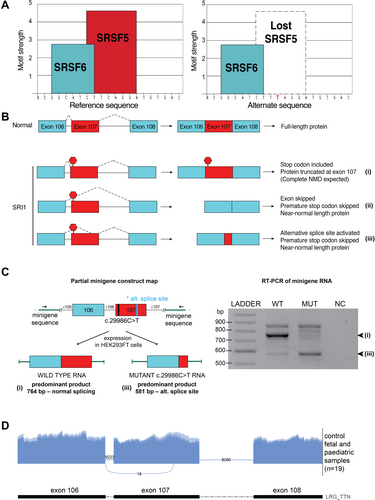
The Family NL1 homozygous frameshift disease-causing variant within exon 140 (PSI 99–100% in fetal or neonatal, pediatric, and adult muscle) was predicted to result in transcripts with reduced inclusion of exon 140, but no change to the reading frame (in-frame exon skipping).
Five additional truncating disease-causing variants were predicted to result in a loss of splicing enhancer elements (n = 3) or gain of heterogeneous nuclear ribonucleoprotein (hnRNP) A1 splicing silencer elements (n = 2) (Table S1A: columns V and W). These would result in skipping of the affected exon and in-frame transcription.
Genotype–Phenotype Correlation
The characteristics of disease-causing variants present in our severe congenital (SC) cohort members (Table S1A) were compared to the characteristics of disease-causing variants present in the 5 adult survivor (AS) cohort members from our previous study to explore possible reasons for the marked differences in clinical presentation (AS cases 1–5 in Table S1B: reported previously as Family 1 [AUS0001], Family 5 [AUS0004], Family 9 [F26], Family 19 [HK0001], and Family 26 [B13-26]).20
- Disease-causing variants in multiple different domains (A-, I-, and M-band), with no group-specific clustering.
- Disease-causing variants in N2A exons (all associated with 100% PSI values across all age groups).
- Disease-causing variants in non-N2A exons associated with variable PSI values.
- Truncating disease-causing variants predominantly predicted to result in complete (rather than incomplete) nonsense mediated decay.
- Splice site disease-causing variants predominantly predicted or shown (via RNA studies) to cause partial or complete exon skipping without frameshift.
- Truncating disease-causing variants predicted to have an additional previously unrecognized splice impact.
Relationship between Cardiac Involvement and Cardiac Isoform Impact
Many affected cohort members died before comprehensive cardiac assessment, or perhaps before manifesting cardiac involvement. Nevertheless, cardiac abnormalities (predominantly septal defects) were present in at least 1 case from 5 of the 11 cohort families (Table S1A: column AA). Two of these 5 families had biallelic N2B/N2BA-impacting disease-causing variants.
Cardiac isoform analyses based on findings in the families from this study cohort and an additional 50 published families with convincingly pathogenic segregation confirmed disease-causing variants (Table S3) show a significant association between cardiac involvement and the presence of 2 N2B/N2BA-impacting disease-causing variants (Fisher's exact test: p-value, 0.018; odds ratio, 3.90; 95% confidence interval, 1.20–13.53).
The mother of AUS2.II.3, who developed atrial fibrillation before the age of 40, was the carrier of a N2B/N2BA-impacting disease-causing variant. Further statistical analysis was not undertaken because of incomplete carrier family member cardiac screening.
Discussion
RT has recently emerged as a common genetic muscle disorder, largely because of the exponential increase in MPS use within the diagnostic setting. The growing number of genetically confirmed cases has facilitated a rapid expansion in our understanding of the clinical spectrum of this disorder, which we now know includes patients with prenatal-, infant-, childhood-, adolescent-, and occasional adult-onset presentations, variable rates of progression, and different complication (eg, cardiac risk) profiles.
At the extreme end of the clinical severity spectrum are congenital titinopathy cases with severe-to-profound prenatal-onset weakness, limb contractures, hydrops, and/or growth restriction, frequently resulting in in utero or early infant death or pregnancy termination because of poor prognosis. At the other end of the spectrum are the more than 20 reported congenital titinopathy cases with convincingly pathogenic disease-causing variants who have survived into adulthood, many of whom remain somewhat ambulant and independent in many activities of daily living.16, 20-25 There are also more than 15 reported non-congenital RT cases (onset after infancy) who have reached adulthood.13, 20, 21, 24, 25, 43 The reasons for such vast differences in clinical severity remain unclear.
Congenital long bone fractures (sometimes multiple) were present in 4 of 9 cohort members who survived until birth. Congenital fractures are rare, but have been reported in association with motor pathway disorders caused by disease-causing variants in a range of disease genes including SMN1 (Type 0 SMA),44 ASCC1,45, 46 BICD2,47 EXOSC9,48 LMOD3,49 KLHL40,50 RYR1,51 and ACTA1.52 The reason why only a small subset of severely-to-profoundly weak infants with these genetic diagnoses present with this complication remains unclear. It is possible that only a small proportion of infants are put at risk of fractures because of their in utero positioning toward the end of pregnancy and/or at time of delivery. Of note, is that 2 of cases had congenital fractures (from AUS1, MAL1) despite having an elective Caesarean section, which should have reduced their fracture risk. Congenital fracture-associated disorders might also have additional yet-to-be-characterized impacts on bone development.
The complete absence of sternocleidomastoid, right biceps, and thigh muscle groups noted at autopsy in 1 member of this cohort (MAL1.II.4) was a rare and unexpected finding. Of additional note, is that extreme fat replacement of quadriceps muscle was noted in 1 additional genetically confirmed congenital titinopathy patient not included in this cohort. In this young infant, no diagnostically useful quadriceps muscle fibers could be obtained via open muscle biopsy. Thigh muscle ultrasound suggested increased echogenicity, but the presence of at least some muscle. A second open muscle biopsy with surgical exploration deeper into the quadriceps femoris muscle fascia yielded fatty tissue and rare muscle fibers with centralized/internalized nuclei (Dr Adnan Manzur, personal communication).
The diagnosis of congenital titinopathy in MAL1.II.4 was established following a neuromuscular panel analysis. Although no pathogenic or likely pathogenic variants in other muscle disease genes were identified by this analysis, it remains possible that additional variants in 1 or more additional muscle disease genes (known, or as-yet-unknown) contributed to this finding. Of additional note, is that this patient's parents were consanguineous. The likelihood of involvement of additional genes responsible for recessive muscle conditions is, therefore, higher than it would be for a patient with non-consanguineous parents. However, an alternative hypothesis is that, in a subset of congenital titinopathy patients, fetal expression of isoforms critical to the development of specific muscle groups are so severely reduced during early prenatal life that muscle development is profoundly compromised, or there is complete failure of muscle development.
The strikingly unusual central and circumferential mitochondrial accumulations noted in 2 cohort member biopsies appear to be more common in biopsies from recessive (including congenital) titinopathy cases than in biopsies from individuals with other muscle disorders53 and may be an important clue to this diagnosis.
In this study, disease-causing variant-containing exons not included within the canonical N2A mature skeletal muscle transcript (non-N2A exons) were associated with moderate to high fetal/neonatal PSI values (56–100%). This finding underscores the importance of non-N2A exons during fetal muscle development. Disease-causing variants in 3 non-N2A exons were present in 4 of our 11 severely affected cohort families, including 1 consanguineous family (MAL1). Disease-causing variants in non-N2A exons have also been reported in numerous published congenital titinopathy cases.20, 21, 24 It is, therefore, clear that disease-causing variants in non-N2A exons can contribute to disease, including severe disease. All TTN exons (not just N2A exons) should, therefore, be interrogated during the diagnostic evaluation of TTN.
Of note, I-band exons 173–199 lie within the TTN triplicated repeat region. RNA-seq reads from this region do not map as accurately as reads from other regions. The in vivo usage of these exons might, therefore, be higher than suggested by our PSI calculations.
In this study, using cohort and additional published data, we were able to confirm that congenital titinopathy patients with convincingly pathogenic biallelic TTN disease-causing variants that impact N2B and N2BA (the 2 most abundant cardiac isoforms), are significantly more likely to develop cardiac complications than cases with other combinations of disease-causing variants. However, because the other genetic and environmental factors that contribute to an increased risk of cardiac involvement have not been fully elucidated, regular cardiac screening is still strongly recommended for all patients and carrier relatives. Cardiac assessment is particularly important for maternal carrier family members who are planning pregnancy or are pregnant because of the well-recognized risk of cardiac decompensation during or shortly after pregnancy.54, 55
As noted in previous studies,20, 26 all congenital titinopathy cases reported to date have had at least 1 disease-causing variant predicted or shown (via western blot) to result in the production of at least some near-normal length titin. Examples of “titin preserving” disease-causing variants include (1) C-terminal M-band truncating variants that do not result in complete nonsense mediated decay, (2) splice-altering variants that result in in-frame loss of a single exon at the transcript level, and (3) disease-causing variants that impact lower PSI exons not included in all transcripts.
In this study, 3 families (SRI1, NL1, and NL2) initially appeared to be exceptions to this rule. However, subsequent in silico analyses of the disease-causing variants present in families in SRI1 and NL1 suggested that the homozygous causative variants in each family had an additional previously unrecognized impact on pre-mRNA splicing.
The additional splicing impact of the Family SRI1 exon 107 nonsense disease-causing variant was subsequently confirmed via minigene assay. Because no patient muscle was available it was not possible to determine the in vivo levels of titin transcript produced by this additional splicing impact. However, in this family, this splicing impact appears to have resulted in sufficient titin expression to support the survival of all 3 affected infants, at least until birth.
In Family NL1, both affected pregnancies were terminated early (11 and 20 weeks). It is, therefore, not clear if these cases would have survived until birth. However, survival to 20 weeks had been possible even though the homozygous exon 140 frameshift disease-causing variant present in these cases impacted a 99 to 100% PSI exon and was predicted to result in complete nonsense-mediated decay. In this family, the additional splice impact is predicted to cause in-frame skipping of exon 140. This may have resulted in sufficient titin expression to support early fetal survival (until 20 weeks gestation).
In Family NL2, 1 of the 2 causative nonsense disease-causing variants (in 100% PSI exons 64 and 135) ware predicted to result in incomplete NMD. This exon 64 nonsense variant may, therefore, have resulted in production of at least some truncated protein, although any protein produced from this allele is likely to have been extremely short. Of note, both affected cases in this family were terminated early (latest at 14 weeks) because of severe abnormalities. Survival beyond this point may not have been possible.
Overall, the disease-causing variant-related findings in these and all other reported congenital titinopathy cases suggest that production of at least some near-normal length titin is essential to early human survival, and that combinations of biallelic TTN disease-causing variants that result in severely reduced or absent titin compromise pregnancy viability. This hypothesis is further supported by studies that have shown that Ttn-deficient mice do not survive embryogenesis.56
Comparison of the predicted transcript- and protein-level impacts of the disease-causing variants present in severe congenital and adult survivor cases was inconclusive. There were no obvious points of difference in terms of disease-causing variant type or location, usage of impacted exons, degree of predicted NMD, or occurrence of previously unrecognized additional splice impacts. This may, in part, be because of the limited size of the comparison groups. All the above-mentioned factors, perhaps in addition to as-yet-uncharacterized transcriptional and translational regulatory factors, likely contribute to overall differences in titin abundance and/or length.
Although not yet proven, our somewhat unexpected findings in SRI1, NL1, and NL2 strongly suggest that differences in titin protein abundance impact clinical outcomes including in utero survival. We hypothesize that differences in titin abundance in patient muscle account for at least some of the differences in severity observed clinically in cases that survive beyond birth.
The length of “remnant titins” expressed in patient muscle might also impact severity. Shorter remnant titins might destabilize the sarcomere, increase vulnerability to sarcomeric wear and tear over time, and/or adversely impact critical signaling pathways. One possibility is that there is a titin length threshold. Titins shorter than this length may critically impair key structural and/or signaling functions. There may also be 1 or more titin domains that when absent, result in more severe clinical presentations. This impact may be exacerbated by reduced overall titin transcript/protein levels and/or significantly shortened remnant proteins.
To explore these possibilities, short- and long-read transcriptomic and protein-based analyses of muscle from patients with a range of different clinical severities is needed. If our hypotheses are confirmed, these types of analyses could facilitate clinical prediction. In addition, these analyses are also likely to inform our understanding of whether increasing titin abundance and/or length might be an effective treatment strategy, and how much additional titin might be needed to achieve significantly improved clinical outcomes.
Acknowledgements
This study is funded by the Belgian Kids' Fund for Pediatric Research, the Fonds Erasme, the Belgian Association Against Neuromuscular Diseases and the Belgian Royal Academy of Medicine (Prize Professor Christian Coërs 2015, S.C.). This study is funded by an Australian NHMRC Neil Hamilton Fairley Early Career Research Fellowship (GNT1090428, E.O.). This study is supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London (F.M.). This study is supported by the Luminesce Alliance - Innovation for Children's Health (P.S., M.P., and M.C.). The support of Muscular Dystrophy UK, MRC and BRC Neuromuscular Centre Biobank and NHS England Highly Specialised Services to the Dubowitz Neuromuscular Centre is gratefully acknowledged. This study received funding from the New South Wales State Government Research Attraction and Acceleration Program (S.C. and M.W.). This study is supported by an Australian NHMRC Fellowship (GNT1122952, G.R.) and Ideas Grant (GNT2002640, G.R.). This study is supported by an Australian NHMRC Investigator Grant (GNT1176265, M.P.).
Author Contributions
S.C., N.D., P.S., A.S., K.J., M.W., R.P., M.D., F.M., and E.O. contributed to the conception and design of the study; S.C., N.D., P.S., A.S., J.C., K.G., C.V., H.K., S.B., F.F., L.W., F.E., M.B., J.P., A.C., S.B., N.G., M.K., C.T., N.F., M.I., N.T., S.E., I.M., S.P., C.F., J.B., G.B., S.H., K.A., D.K., G.B., B.W., M.W., K.V., A.S., A.M., A.M., I.S., M.S., C.M., D.S., K.S., T.G., Y.A., S.C., L.R., H.J., E.K., A.M., S.C., G.R., M.C., M.P., the Titin Research Consortium, R.P., M.D., F.M., and E.O. contributed to the acquisition and analysis of data; S.C., N.D., A.S., K.J., S.C., N.L., H.J., G.R., M.W., M.C., M.P., F.M., and E.O. contributed to drafting the text or preparing the figures.
Potential Conflicts of Interest
Nothing to report.
Appendix A: Titin Research Consortium
Ana Ferreiro MD, PhD, Isabelle Richard PhD, Carsten G. Bönnemann MD, Sandra Donkervoort MS, CGC, Ho-Ming Luk MD, FRCPCH (UK), Hugo Sampaio MBBCh, FRACP, MPhil, Michelle A. Farrar MBBS, FRACP, PhD, David Mowat MBBS, Robin B. Fitzsimons MBBS, BSc(Med), PhD, FRACP, Carole Vuillerot MD, PhD.
Open Research
Data Availability
A comprehensive summary of disease-causing variant data is provided in Table S1. All described members of this cohort had an established genetic diagnosis of congenital titinopathy (biallelic disease-causing TTN variants) before study inclusion. Primary whole exome sequencing data, whole genome sequencing data, epigenetic data, and microarray data were not generated during this study.