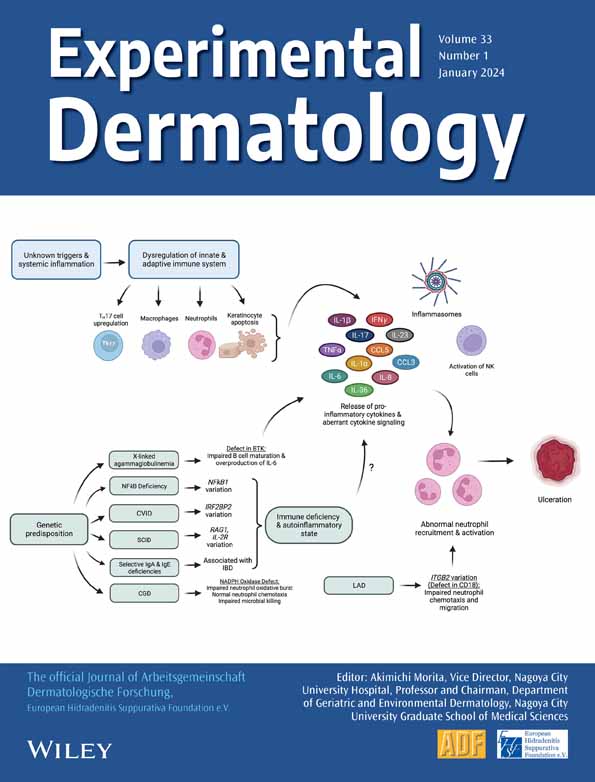
NOD2 at the interface of hidradenitis suppurativa and inflammatory bowel disease—An in silico analysis
Dillon Mintoff
Department of Pathology, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Department of Dermatology, Mater Dei Hospital, Msida, Malta
Search for more papers by this authorIsabella Borg
Department of Pathology, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Department of Pathology, Section of Clinical Genetics, Mater Dei Hospital, Msida, Malta
Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta
Search for more papers by this authorCorresponding Author
Nikolai P. Pace
Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta
Department of Anatomy, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Correspondence
Nikolai P. Pace, Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta.
Email: [email protected]
Search for more papers by this authorDillon Mintoff
Department of Pathology, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Department of Dermatology, Mater Dei Hospital, Msida, Malta
Search for more papers by this authorIsabella Borg
Department of Pathology, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Department of Pathology, Section of Clinical Genetics, Mater Dei Hospital, Msida, Malta
Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta
Search for more papers by this authorCorresponding Author
Nikolai P. Pace
Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta
Department of Anatomy, Faculty of Medicine and Surgery, University of Malta, Msida, Malta
Correspondence
Nikolai P. Pace, Centre for Molecular Medicine and Biobanking, University of Malta, Msida, Malta.
Email: [email protected]
Search for more papers by this authorOpen Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in GnomAD at https://gnomad.broadinstitute.org/. These data were derived from the following resources available in the public domain: PubMed, https://gnomad.broadinstitute.org/.
Supporting Information
| Filename | Description |
|---|---|
| exd14928-sup-0001-TableS1.docxWord 2007 document , 29.2 KB |
Table S1: Summary of NOD2 variants identified in patients with hidradenitis suppurativa. Variant classification was interpreted using Varsome6 and Franklin (https://franklin.genoox.com). The impact of coding variants on protein structural conformation, stability and flexibility was predicted using DynaMut2,7 and Missense3D8 using the Alpha Fold Predicted Structure9 (accession code Q8K3Z0). Tolerance to variation was assessed using the MetaDome webserver.4 Further in silico prediction was carried out using MetaRNN, a recurrent neural network based ensemble prediction score, which incorporates 16 functional prediction scores (including SIFT), 8 conservation scores and allele frequency information. ACMG, American College of Medical Genetics; IBD, inflammatory bowel disease; PASH, pyoderma gangrenosum, acne, and hidradenitis suppurativa. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1Pace NP, Mintoff D, Borg I. The genomic architecture of hidradenitis suppurativa—a systematic review. Front Genet. 2022; 13:861241. doi:10.3389/fgene.2022.861241
- 2Mintoff D, Pace NP, Borg I. Interpreting the spectrum of gamma-secretase complex missense variation in the context of hidradenitis suppurativa-an in-silico study. Front Genet. 2022; 13:962449. doi:10.3389/fgene.2022.962449
- 3Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annu Rev Med. 2007; 58: 401-416. doi:10.1146/annurev.med.58.061705.145024
- 4Li C, Zhi D, Wang K, Liu X. MetaRNN: differentiating rare pathogenic and rare benign missense SNVs and InDels using deep learning. Genome Med. 2022; 14(1): 115. doi:10.1186/s13073-022-01120-z
- 5Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001; 411(6837): 599-603. doi:10.1038/35079107
- 6Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C. MetaDome: pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. 2019; 40(8): 1030-1038. doi:10.1002/humu.23798
- 7Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017; 100(6): 895-906. doi:10.1016/j.ajhg.2017.04.015
- 8Gambichler T, Hessam S, Skrygan M, Bakirtzi M, Kasakovski D, Bechara FG. NOD2 signalling in hidradenitis suppurativa. Clin Exp Dermatol. 2021; 46(8): 1488-1494. doi:10.1111/ced.14773
- 9Sidiq T, Yoshihama S, Downs I, Kobayashi KS. Nod2: a critical regulator of Ileal microbiota and Crohn's disease. Front Immunol. 2016; 7:367. doi:10.3389/fimmu.2016.00367
- 10Ring HC, Thorsen J, Lilje B, et al. Predictive metagenomic analysis identifies specific bacterial metabolic pathways in hidradenitis suppurativa tunnels. J Eur Acad Dermatol Venereol. 2023. doi:10.1111/jdv.19433