Saliva biopsy: Detecting the difference of EBV DNA methylation in the diagnosis of nasopharyngeal carcinoma
Xiao-Hui Zheng
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorChang-Mi Deng
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorTing Zhou
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorXi-Zhao Li
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCao-Li Tang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCheng-Tao Jiang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorYing Liao
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorTong-Min Wang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorYong-Qiao He
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCorresponding Author
Wei-Hua Jia
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Correspondence
Wei-Hua Jia, Sun Yat-Sen University Cancer Center, 651 Dongfeng East Road, Guangzhou, Guangdong 510060, China.
Email: [email protected]
Search for more papers by this authorXiao-Hui Zheng
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorChang-Mi Deng
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorTing Zhou
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorXi-Zhao Li
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCao-Li Tang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCheng-Tao Jiang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorYing Liao
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorTong-Min Wang
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorYong-Qiao He
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Search for more papers by this authorCorresponding Author
Wei-Hua Jia
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-Sen University Cancer Center, Guangzhou, Guangdong, China
Correspondence
Wei-Hua Jia, Sun Yat-Sen University Cancer Center, 651 Dongfeng East Road, Guangzhou, Guangdong 510060, China.
Email: [email protected]
Search for more papers by this authorXiao-Hui Zheng and Chang-Mi Deng have contributed equally to this study.
Abstract
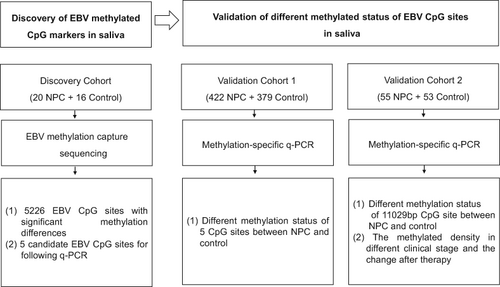
Saliva sampling is a non-invasive method, and could be performed by donors themselves. However, there are few studies reporting biomarkers in saliva in the diagnosis of NPC. A total of 987 salivary samples were used in this study. First, EBV DNA methylation was profiled by capture sequencing in the discovery cohort (n = 36). Second, a q-PCR based method was developed and five representative EBV DNA CpG sites (11 029 bp, 45 849 bp, 57 945 bp, 66 226 bp and 128 102 bp) were selected and quantified to obtain the methylated density in the validation cohort1 (n = 801). Third, a validation cohort2 (n = 108) was used to further verify the differences of EBV methylation in saliva. A significant increase of EBV methylation was found in NPC patients compared with controls. The methylated score of EBV genome obtained by capture sequencing could distinguish NPC from controls (sensitivity 90%, specificity 100%). Further, the methylated density of EBV DNA CpG sites revealed by q-PCR showed a good diagnostic performance. The sensitivity and specificity of detecting a single CpG site (11 029 bp) could reach 75.4% and 99.7% in the validation cohort1, and 78.2% and 100% in the validation cohort2. Besides, the methylated density of the CpG site was found to decrease below the COV in NPC patients after therapy, and increase above the COV after recurrence. Our study provides an appealing alternative for the non-invasive detection of NPC without clinical setting. It paves the way for conducting a home-based large-scale screening in the future.
Graphical Abstract
What's new?
While various Epstein-Barr virus (EBV)-related biomarkers have been established as potential screening biomarkers for nasopharyngeal carcinoma, most of them rely on blood or nasopharyngeal swab samples. Using saliva samples, this study found a significant increase in EBV DNA methylation in nasopharyngeal carcinoma patients compared to controls. Detection at a single CpG site could reach a sensitivity of 75.8% and specificity of 99.7%. The methylated density of the CpG site was found to decrease below the cut-off value after therapy and increase above the cut-off value after recurrence. This study potentially provides a non-invasive alternative for the detection of nasopharyngeal carcinoma.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Supporting Information
| Filename | Description |
|---|---|
| ijc34561-sup-0001-Supinfo.pdfPDF document, 615.2 KB | DATA S1. Supporting Information. |
| ijc34561-sup-0002-Tables.xlsExcel spreadsheet, 9.8 MB | TABLE S4. The methylated density of all CpG sites detected in the capture sequencing was listed. TABLE S5. The methylated region divided by 200 bp length and the methylated score was listed TABLE S6. The methylated density of EBV genes divided into four infection phase was listed. TABLE S7. The methylated density of 17 CpG sites located in the EBV DNA was listed. A model containing these 17 CpG sites was used. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2018; 68(6): 394-424.
- 2Lee AWM, Ng WT, Chan LLK, et al. Evolution of treatment for nasopharyngeal cancer: success and setback in the intensity-modulated radiotherapy era. Radiother Oncol. 2014; 110(3): 377-384.
- 3Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004; 4(10): 757-768.
- 4Tsao SW, Yip YL, Tsang CM, et al. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014; 50(5): 330-338.
- 5Wu L, Li C, Pan L. Nasopharyngeal carcinoma: a review of current updates. Exp Ther Med. 2018; 15(4): 3687-3692.
- 6Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet (London, England). 2019; 394(10192): 64-80.
- 7Chien YC, Chen JY, Liu MY, et al. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N Engl J Med. 2001; 345(26): 1877-1882.
- 8Cao SM, Liu Z, Jia WH, et al. Fluctuations of epstein-barr virus serological antibodies and risk for nasopharyngeal carcinoma: a prospective screening study with a 20-year follow-up. PLoS One. 2011; 6(4):e19100.
- 9Liu Y, Huang QH, Liu WL, et al. Establishment of VCA and EBNA1 IgA-based combination by enzyme-linked immunosorbent assay as preferred screening method for nasopharyngeal carcinoma: a two-stage design with a preliminary performance study and a mass screening in southern China. Int J Cancer. 2012; 131(2): 406-416.
- 10Mutirangura A1PW, Theamboonlers A, Sriuranpong V, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998; 4: 665-669.
- 11Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999; 59(6): 1188-1191.
- 12Chan KC, Lo YM. Circulating EBV DNA as a tumor marker for nasopharyngeal carcinoma. Semin Cancer Biol. 2002; 12(6): 489-496.
- 13Yip TTC, Ngan RKC, Fong AHW, Law SCK. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014; 50(6): 527-538.
- 14Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. New Engl J Med. 2017; 377(6): 513-522.
- 15Tune CE, Liavaag PG, Freeman JL, et al. Nasopharyngeal brush biopsies and detection of nasopharyngeal cancer in a high-risk population. J Natl Cancer Inst. 1999; 91(9): 796-800.
- 16Tong JHM, Tsang RKY, Lo KW, et al. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res. 2002; 8(8): 2612-2619.
- 17Hao SP, Tsang NM, Chang KP. Screening nasopharyngeal carcinoma by detection of the latent membrane protein 1 (LMP-1) gene with nasopharyngeal swabs. Cancer. 2003; 97(8): 1909-1913.
- 18Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int J Cancer. 2006; 119(3): 608-614.
- 19Zheng XH, Lu LX, Li XZ, Jia WH. Quantification of Epstein-Barr virus DNA load in nasopharyngeal brushing samples in the diagnosis of nasopharyngeal carcinoma in southern China. Cancer Sci. 2015; 106(9): 1196-1201.
- 20Chen GH, Liu ZW, Yu KJ, et al. Utility of Epstein-Barr virus DNA in nasopharynx swabs as a reflex test to triage seropositive individuals in nasopharyngeal carcinoma screening programs. Clin Chem. 2022; 68(7): 953-962.
- 21Zhang GH, Zong JF, Lin SJ, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015; 136(5): E301-E312.
- 22Jiang C, Chen JN, Xie SH, et al. Evaluation of circulating EBV microRNA BART2-5p in facilitating early detection and screening of nasopharyngeal carcinoma. Int J Cancer. 2018; 143(12): 3209-3217.
- 23Lin C, Lin KY, Zhang B, et al. Plasma Epstein-Barr virus MicroRNA BART8-3p as a diagnostic and prognostic biomarker in nasopharyngeal carcinoma. Oncologist. 2022; 27(4): E340-E349.
- 24Chang HW, Chan A, Kwong DLW, Wei WI, Sham JST, Yuen APW. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int J Cancer. 2003; 105(6): 851-855.
- 25Nawaz I, Moumad K, Martorelli D, et al. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assay. Clin Epigenet. 2015; 7: 7.
- 26Tan GW, Sivanesan VM, Rahman FIA, et al. A novel and non-invasive approach utilising nasal washings for the detection of nasopharyngeal carcinoma. Int J Cancer. 2019; 145(8): 2260-2266.
- 27Liu LZ, Zuo LL, Yang J, et al. Exosomal cyclophilin a as a novel noninvasive biomarker for Epstein-Barr virus associated nasopharyngeal carcinoma. Cancer Med-US. 2019; 8(6): 3142-3151.
- 28Ramayanti O, Verkuijlen SAWM, Novianti P, et al. Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections. Int J Cancer. 2019; 144(10): 2555-2566.
- 29Zou JH, Xiao ZS, Wu Y, Yang JY, Cui N. Noninvasive fecal testing for colorectal cancer. Clin Chim Acta. 2022; 524: 123-131.
- 30Guo R, Gewurz BE. Epigenetic control of the Epstein-Barr lifecycle. Curr Opin Virol. 2022; 52: 78-88.
- 31Fernandez AF, Rosales C, Lopez-Nieva P, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009; 19(3): 438-451.
- 32Ye WM, Chang ET, Liu ZW, et al. Development of a population-based cancer case-control study in southern China. Oncotarget. 2017; 8(50): 87073-87085.
- 33Chen Y, Chang ET, Liu Q, et al. Environmental factors for Epstein-Barr virus reactivation in a high-risk area of nasopharyngeal carcinoma: a population-based study. Open Forum Infect Dis. 2022; 9(5): ofac128.
- 34Xue WQ, He YQ, Liao XY, et al. Decreased oral Epstein-Barr virus DNA loads in patients with nasopharyngeal carcinoma in southern China: a case-control and a family-based study. Cancer Med. 2018; 7(7): 3453-3464.
- 35He YQ, Liao XY, Xue WQ, et al. Association between environmental factors and oral Epstein-Barr virus DNA loads: a multicenter cross-sectional study in China. J Infect Dis. 2019; 219(3): 400-409.
- 36Zhou T, Yang DW, He YQ, et al. Associations between environmental factors and serological Epstein-Barr virus antibodies in patients with nasopharyngeal carcinoma in South China. Cancer Med. 2019; 8(10): 4852-4866.
- 37Lam WKJ, Jiang P, Chan KCA, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun. 2019; 10(1): 3256.
- 38Luo HY, Zhao Q, Wei W, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020; 12(524): eaax7533.
- 39Faulkner GC, Krajewski AS, Crawford DH. The ins and outs of EBV infection. Trends Microbiol. 2000; 8(4): 185-189.
- 40Hammerschmidt W. The epigenetic life cycle of Epstein-Barr virus. Curr Top Microbiol. 2015; 390: 103-117.
- 41Woellmer A, Hammerschmidt W. Epstein-Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr Opin Virol. 2013; 3(3): 260-265.
- 42Lin Z, Swan K, Zhang X, et al. Secreted oral epithelial cell membrane vesicles induce Epstein-Barr virus reactivation in latently infected B cells. J Virol. 2016; 90(7): 3469-3479.
- 43Midoen YH, Suryandari DA, Yunaini L, Susworo R, Auerkari EI, Freisleben HJ. Epstein-Barr virus nuclear antigen-1 is useful as therapeutic efficacy marker in serum but not in saliva of nasopharyngeal cancer patients who underwent radiotherapy. Ecancermedicalscience. 2021; 15: 1254.
- 44Ganapathi R, Kumar RR, Thomas KC, et al. Epstein-Barr virus dynamics and its prognostic impact on nasopharyngeal cancers in a non-endemic region. Ecancermedicalscience. 2022; 16: 1479.
- 45Fung SYH, Lam JWK, Chan KCA. Clinical utility of circulating Epstein-Barr virus DNA analysis for the management of nasopharyngeal carcinoma. Chin Clin Oncol. 2016; 5(2): 18.