DNA Synthesis and Assembly Technologies: From Oligonucleotides to Complete Genomes
Wei Tan
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorXuemei Jia
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorYing Liu
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorCorresponding Author
Chi Yao
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
E-mail: [email protected]Search for more papers by this authorDayong Yang
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
Search for more papers by this authorWei Tan
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorXuemei Jia
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorYing Liu
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
These authors contributed equally.
Search for more papers by this authorCorresponding Author
Chi Yao
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
E-mail: [email protected]Search for more papers by this authorDayong Yang
Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (MOE), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300350 China
Zhejiang Institute of Tianjin University, Ningbo, Zhejiang, 315201 China
Search for more papers by this authorAbstract
Comprehensive Summary
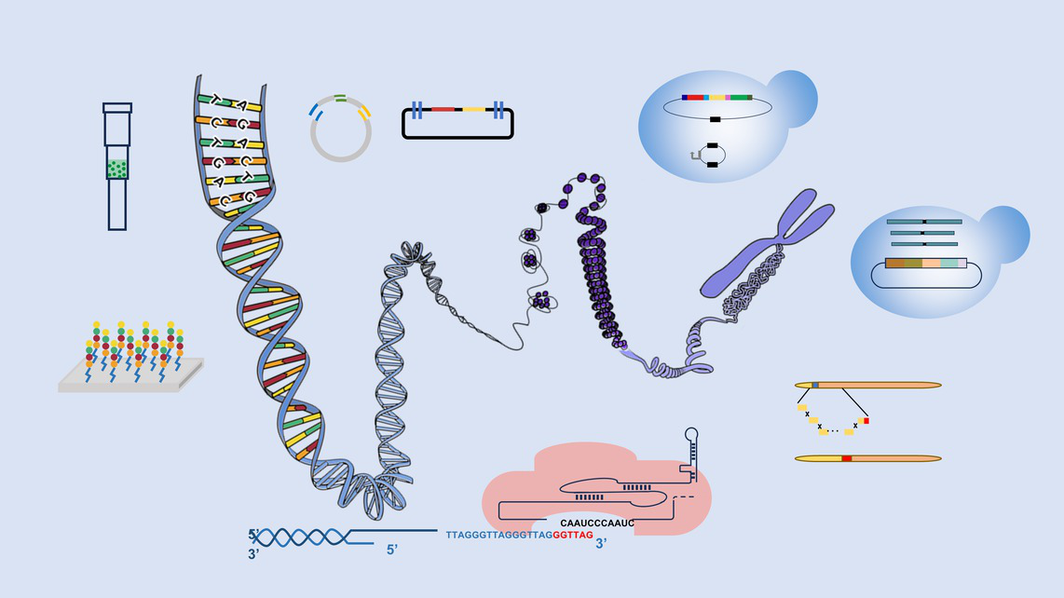
DNA synthesis and assembly technology is the fundamental enabling technology of synthetic biology, providing methods for humans to understand and modify organisms. Oligonucleotide chains and long DNA fragments synthesized through DNA synthesis and assembly technology are becoming increasingly widely used in fields such as biomedicine, energy, new materials, and information storage, with strong application prospects. This review provides a comprehensive and systematic introduction and exposition of current DNA synthesis and assembly technologies, discussing current challenges and future research prospects.
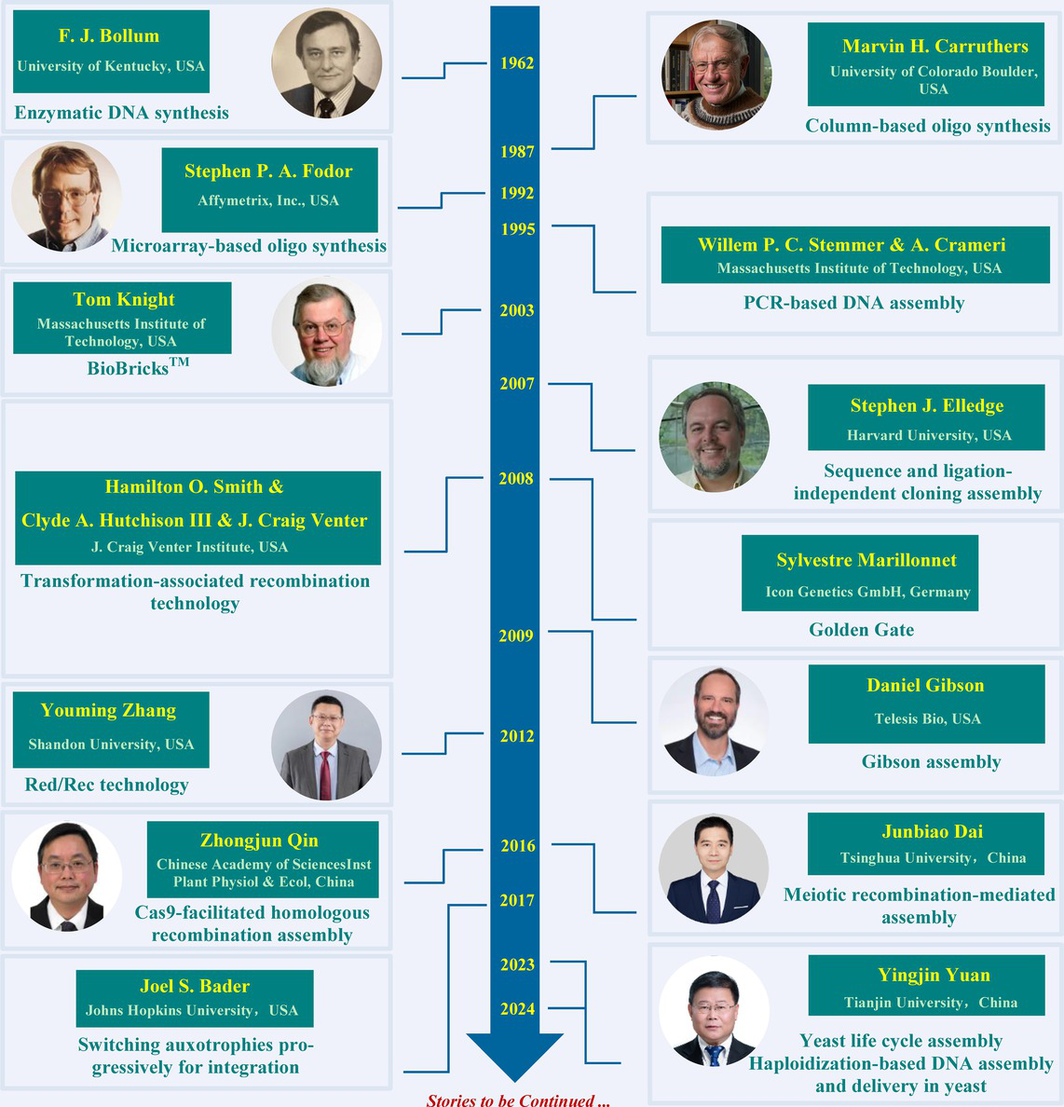
Key Scientists
DNA synthesis and assembly technology is a cutting-edge technology for human understanding and modification of living organisms, with wide applications in the fields of biomedicine, energy, new materials, and information storage. In 1962, the Bollum group first proposed that TDT enzyme could be used for the synthesis of DNA oligonucleotide chains. In 1987, the Carruthers group established column-based oligo synthesis. In 1992, the Fodor group established microarray-based oligo synthesis. In 1995, the Stemmer and Crameri groups established PCR based DNA assembly. In 2003, the Knight group proposed BioBricksTM. In 2007, the Elledge group was the first to report sequence and ligation-independent cloning assembly. In 2008, three scientists Hamilton O. Smith, Clyde A. Hutchison III, and J Craig Venter co-established transformation associated recombination technology. In the same year, the Marillonnet group established Gloden Gate technology. In 2009, the Gibson group established Gibson assembly technology. In 2012, the Zhang group established Red/Rec technology. In 2016, the Qin group established Cas9-facilitated homologous recombination assembly. In the same year, the Dai group established Meiotic recombination-mediated assembly. In 2017, the Bader group established switching auxotrophies progressively for integration. The Yuan group established yeast life cycle assembly in 2023 and established haploidization-based DNA assembly and delivery in yeast in 2024. This review systematically introduces the latest research progress in DNA synthesis and assembly technology, providing a reference for subsequent research.
References
- 1 Khalil, A. S.; Collins, J. J. Synthetic Biology: Applications Come of Age. Nat. Rev. Genet. 2010, 11, 367–379.
- 2 Cameron, D. E.; Bashor, C. J.; Collins, J. J. A Brief History of Synthetic Biology. Nat. Rev. Microbiol. 2014, 12, 381–390.
- 3 Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R. E., Zhao, H. Activation and Characterization of a Cryptic Polycyclic Tetramate Macrolactam Biosynthetic Gene Cluster. Nat. Commun. 2013, 4, 2894.
- 4 Blake, W. J.; Chapman, B. A.; Zindal, A.; Lee, M. E.; Lippow, S. M.; Baynes, B. M. Pairwise Selection Assembly for Sequence-Independent Construction of Long-Length DNA. Nucleic Acids Res. 2010, 38, 2594–2602.
- 5 Hoose, A.; Vellacott, R.; Storch, M.; Freemont, P. S.; Ryadnov, M. G. DNA synthesis technologies to close the gene writing gap. Nat. Rev. Chem. 2023, 7, 144–161.
- 6 Medema, M. H.; Breitling, R.; Bovenberg, R.; Takano, E. Exploiting Plug-and-Play Synthetic Biology for Drug Discovery and Production in Microorganisms. Nat. Rev. Microbiol. 2011, 9, 131–137.
- 7 Gibson, D. G. Synthesis of DNA Fragments in Yeast by One-Step Assembly of Overlapping Oligonucleotides. Nucleic Acids Res. 2009, 37, 6984–6990.
- 8 Jiang, X.; Wang, Y.; Shen, Y. The Review of DNA Synthesis Technologies and Instruments Development. J. Integr. Technol. 2021, 10, 80–95.
- 9 Lin, Q.; Jia, B.; Mitchell, L. A.; Luo, J.; Yang, K.; Zeller, K. I.; Zhang, W.; Xu, Z.; Stracquadanio, G.; Bader, J. S.; Boeke, J. D.; Yuan, Y. J. RADOM, an Efficient in vivo Method for Assembling Designed DNA Fragments Up to 10 kb Long in Saccharomyces Cerevisiae. ACS Synth. Biol. 2015, 4, 213–220.
- 10 Juhas, M.; Ajioka, J. W. High Molecular Weight DNA Assembly in vivo for Synthetic Biology Applications. Crit. Rev. Biotechnol. 2017, 37, 277–286.
- 11 Ellis, T.; Adie, T.; Baldwin, G. S. DNA Assembly for Synthetic Biology: From Parts to Pathways and Beyond. Integr. Biol. 2011, 3, 109–118.
- 12 Michelson, A. M.; Todd, A. R. Nucleotides Part XXXII. Synthesis of a Dithymidine Dinucleotide Containing a 3' : 5'-Internucleotidic Linkage. J. Chem. Soc. 1955, 0, 2632–2638.
- 13 Matteucci, M. D.; Caruthers, M. H. Synthesis of Deoxyoligonucleotides on a Polymer Support. J. Am. Chem. Soc. 1981, 103, 3185–3191.
- 14 Beaucage, S. L.; Caruthers, M. H. Deoxynucleoside Phosphoramidites—A New Class of Key Intermediates for Deoxypolynucleotide Synthesis. Tetrahedron Lett. 1981, 22, 1859–1862.
- 15 Kosuri, S.; Church, G. M. Large-Scale De Novo DNA Synthesis: Technologies and Applications. Nat. Methods 2014, 11, 499–507.
- 16 Kozlov, I. A.; Dang, M.; Sikes, K.; Kotseroglou, T.; Barker, D. L.; Zhao, C. Significant Improvement of Quality for Long Oligonucleotides by Using Controlled Pore Glass with Large Pores. Nucleos. Nucleot. Nucl. 2005, 24, 1037–1041.
- 17 LeProust, E. M.; Peck, B. J.; Spirin, K.; McCuen, H. B.; Moore, B.; Namsaraev, E.; Caruthers, M. H. Synthesis of High-Quality Libraries of Long (150mer) Oligonucleotides by a Novel Depurination Controlled Process. Nucleic Acids Res. 2010, 38, 2522–2540.
- 18 Jensen, M.; Roberts, L.; Johnson, A.; Fukushima, M.; Davis, R. Next Generation 1536-Well Oligonucleotide Synthesizer with On-The-Fly Dispense. J. Biotechnol. 2014, 171, 76–81.
- 19 Deshpande, S.; Yang, Y.; Chilkoti, A.; Zauscher, S. Enzymatic Synthesis and Modification of High Molecular Weight DNA Using Terminal Deoxynucleotidyl Transferase. Method. Enzymol. 2019, 627, 163–188.
- 20 Ma, S.; Saaem, I.; Tian, J. Error Correction in Gene Synthesis Technology. Trends Biotechnol. 2012, 30, 147–154.
- 21 Levrie, K.; Jans, K.; Schepers, G.; Vos, R.; Van Dorpe, P.; Lagae, L.; Van Hoof, C.; Van Aerschot, A.; Stakenborg, T. Direct On-Chip DNA Synthesis Using Electrochemically Modified Gold Electrodes as Solid Support. Jpn. J. Appl. Phys. 2018, 57, 04fm01.
- 22 Gao, X.; Yu, P.; LeProust, E.; Sonigo, L.; Pellois, J. P.; Zhang, H. Oligonucleotide Synthesis Using Solution Photogenerated Acids. J. Am. Chem. Soc. 1998, 120, 12698–12699.
- 23 Maurer, K.; Cooper, J.; Caraballo, M.; Crye, J.; Suciu, D.; Ghindilis, A.; Leonetti, J. A.; Wang, W.; Rossi, F. M.; Stover, A. G.; Larson, C.; Gao, H.; Dill, K.; McShea, A. Electrochemically Generated Acid and Its Containment to 100 Micron Reaction Areas for the Production of DNA Microarrays. PLoS One 2006, 1, e34.
- 24 Hughes, T. R.; Mao, M.; Jones, A. R.; Burchard, J.; Marton, M. J.; Shannon, K. W.; Lefkowitz, S. M.; Ziman, M.; Schelter, J. M.; Meyer, M. R.; Kobayashi, S.; Davis, C.; Dai, H.; He, Y. D.; Stephaniants, S. B.; Cavet, G.; Walker, W. L.; West, A.; Coffey, E.; Shoemaker, D. D.; Stoughton, R.; Blanchard, A. P.; Friend, S. H.; Linsley, P. S. Expression Profiling Using Microarrays Fabricated by an Ink-Jet Oligonucleotide Synthesizer. Nat. Biotechnol. 2001, 19, 342–347.
- 25 Huang, X.; Dai, J. DNA Synthesis Technology: Foundation of DNA Data Storage. Syn. Biol. J. 2021, 2, 335–353.
- 26 Palluk, S.; Arlow, D. H.; de Rond, T.; Barthel, S.; Kang, J. S.; Bector, R.; Baghdassarian, H. M.; Truong, A. N.; Kim, P. W.; Singh, A. K.; Hillson, N. J.; Keasling, J. D. De Novo DNA Synthesis Using Polymerase-Nucleotide Conjugates. Nat. Biotechnol. 2018, 36, 645–650.
- 27 Eisenstein, M. Enzymatic DNA Synthesis Enters New Phase. Nat. Biotechnol. 2020, 38, 1113–1115.
- 28 Jensen, M. A.; Davis, R. W. Template-Independent Enzymatic Oligonucleotide Synthesis (TiEOS): Its History, Prospects, and Challenges. Biochemistry 2018, 57, 1821–1832.
- 29 Bollum, F. J. Thermal Conversion of Nonpriming Deoxyribonucleic Acid to Primer. J. Biol. Chem. 1959, 234, 2733–2734.
- 30 Tjong, V.; Yu, H.; Hucknall, A.; Rangarajan, S.; Chilkoti, A. Amplified On-Chip Fluorescence Detection of DNA Hybridization by Surface-Initiated Enzymatic Polymerization. Anal. Chem. 2011, 83, 5153–5159.
- 31 Mathews, A. S.; Yang, H.; Montemagno, C. Photo-Cleavable Nucleotides for Primer Free Enzyme Mediated DNA Synthesis. Org. Biomol. Chem. 2016, 14, 8278–8288.
- 32 Lee, H.; Wiegand, D. J.; Griswold, K.; Punthambaker, S.; Chun, H.; Kohman, R. E.; Church, G. M. Photon-Directed Multiplexed Enzymatic DNA Synthesis for Molecular Digital Data Storage. Nat. Commun. 2020, 11, 5246.
- 33 Lu, X.; Li, J.; Li, C.; Lou, Q.; Peng, K.; Cai, B.; Liu, Y.; Yao, Y.; Lu, L.; Tian, Z.; Ma, H.; Wang, W.; Cheng, J.; Guo, X.; Jiang, H.; Ma, Y. Enzymatic DNA Synthesis by Engineering Terminal Deoxynucleotidyl Transferase. ACS Catal. 2022, 12, 2988–2997.
- 34 Kodumal, S. J.; Patel, K. G.; Reid, R.; Menzella, H. G.; Welch, M.; Santi, D. V. Total Synthesis of Long DNA Sequences: Synthesis of a Contiguous 32-kb Polyketide Synthase Gene Cluster. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15573–15578.
- 35 Goffeau, A.; Barrell, B. G.; Bussey, H.; Davis, R. W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J. D.; Jacq, C.; Johnston, M.; Louis, E. J.; Mewes, H. W.; Murakami, Y.; Philippsen, P.; Tettelin, H.; Oliver, S. G. Life with 6000 Genes. Science 1996, 274, 563–567.
- 36 Sanger, F.; Nicklen, S.; Coulson, A. R. DNA Sequencing with Chain Terminating Inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977, 74, 5463–5467.
- 37 Mahmoud, M.; Huang, Y.; Garimella, K.; Audano, P. A.; Wan, W.; Prasad, N.; Handsaker, R. E.; Hall, S.; Pionzio, A.; Schatz, M. C.; Talkowski, M. E.; Eichler, E. E.; Levy, S. E.; Sedlazeck, F. J. Utility of Long-Read Sequencing for All of Us. Nat. Commun. 2024, 15, 837.
- 38 Jain, M.; Olsen, H. E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 239.
- 39 Shao, Z.; Luo, Y.; Zhao, H. DNA Assembler Method for Construction of Zeaxanthin-Producing Strains of Saccharomyces Cerevisiae. Methods Mol. Biol. 2012, 898, 251–262.
- 40 Ma, S.; Tang, N.; Tian, J. DNA Synthesis, Assembly and Applications in Synthetic Biology. Curr. Opin. Chem. Biol. 2012, 16, 260–267.
- 41 Chang, H.; Wang, C.; Wang, P.; Zhou, J.; Li, B. DNA Assembly Technologies: A Review. Chin. J. Biotech. 2019, 35, 2215–2226.
- 42 Clarke, L.; Kitney, R. Developing Synthetic Biology for Industrial Biotechnology Applications. Biochem. Soc. T. 2020, 48, 113–122.
- 43 Gibson, D. G.; Young, L.; Chuang, R. Y.; Venter, J. C.; Hutchison, C. A., 3rd; Smith, H. O. Enzymatic Assembly of DNA Molecules Up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345.
- 44 Gibson, D. G.; Glass, J. I.; Lartigue, C.; Noskov, V. N.; Chuang, R. Y.; Algire, M. A.; Benders, G. A.; Montague, M. G.; Ma, L.; Moodie, M. M.; Merryman, C.; Vashee, S.; Krishnakumar, R.; Assad-Garcia, N.; Andrews-Pfannkoch, C.; Denisova, E. A.; Young, L.; Qi, Z. Q.; Segall-Shapiro, T. H.; Calvey, C. H.; Parmar, P. P.; Hutchison, C. A., 3rd; Smith, H. O.; Venter, J. C. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 2010, 329, 52–56.
- 45 Esvelt, K. M.; Mali, P.; Braff, J. L.; Moosburner, M.; Yaung, S. J.; Church, G. M. Orthogonal Cas9 Proteins for RNA-Guided Gene Regulation and Editing. Nat. Methods 2013, 10, 1116–1121.
- 46 Hutchison, C. A., 3rd; Chuang, R. Y.; Noskov, V. N.; Assad-Garcia, N.; Deerinck, T. J.; Ellisman, M. H.; Gill, J.; Kannan, K.; Karas, B. J.; Ma, L.; Pelletier, J. F.; Qi, Z. Q.; Richter, R. A.; Strychalski, E. A.; Sun, L.; Suzuki, Y.; Tsvetanova, B.; Wise, K. S.; Smith, H. O.; Glass, J. I.; Merryman, C.; Gibson, D. G.; Venter, J. C. Design and Synthesis of a Minimal Bacterial Genome. Science 2016, 351, aad6253.
- 47 Engler, C.; Gruetzner, R.; Kandzia, R.; Marillonnet, S. Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS One 2009, 4, e5553.
- 48 Cermak, T.; Doyle, E. L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J. A.; Somia, N. V.; Bogdanove, A. J.; Voytas, D. F. Efficient Design and Assembly of Custom TALEN and Other TAL Effector-Based Constructs for DNA Targeting. Nucleic Acids Res. 2011, 39, e82.
- 49 Lauressergues, D.; Couzigou, J. M.; Clemente, H. S.; Martinez, Y.; Dunand, C.; Becard, G.; Combier, J. P. Primary Transcripts of MicroRNAs Encode Regulatory Peptides. Nature 2015, 520, 90–93.
- 50 Goosens, V. J.; Walker, K. T.; Aragon, S. M.; Singh, A.; Senthivel, V. R.; Dekker, L.; Caro-Astorga, J.; Buat, M. L. A.; Song, W.; Lee, K. Y.; Ellis, T. Komagataeibacter Tool Kit (KTK): a Modular Cloning System for Multigene Constructs and Programmed Protein Secretion from Cellulose Producing Bacteria. ACS Synth. Biol. 2021, 10, 3422–3434.
- 51 Cepleanu-Pascu, I. A.; Stan, M.; Cocioba, S.; Stoica, I. Easy Method for Six-Fragment Golden Gate Assembly of Modular Vectors. Biotechniques 2023, 74, 85–99.
- 52 Sleight, S. C.; Bartley, B. A.; Lieviant, J. A.; Sauro, H. M. In-Fusion BioBrick Assembly and Re-Engineering. Nucleic Acids Res. 2010, 38, 2624–2636.
- 53 Liu, J. K.; Chen, W. H.; Ren, S. X.; Zhao, G. P.; Wang, J. iBrick: A New Standard for Iterative Assembly of Biological Parts with Homing Endonucleases. PLoS One 2014, 9, e110852.
- 54 Gibson, D. G.; Benders, G. A.; Axelrod, K. C.; Zaveri, J.; Algire, M. A.; Moodie, M.; Montague, M. G.; Venter, J. C.; Smith, H. O.; Hutchison, C. A., 3rd. One-Step Assembly in Yeast of 25 Overlapping DNA Fragments to Form a Complete Synthetic Mycoplasma Genitalium Genome. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 20404–20409.
- 55 Warrens, A. N.; Jones, M. D.; Lechler, R. I. Splicing by Overlap Extension by PCR Using Asymmetric Amplification: an Improved Technique for the Generation of Hybrid Proteins of Immunological Interest. Gene 1997, 186, 29–35.
- 56 Quan, J.; Tian, J. Circular Polymerase Extension Cloning of Complex Gene Libraries and Pathways. PLoS One 2009, 4, e6441.
- 57 Li, M. Z.; Elledge, S. J. SLIC: a Method for Sequence- and Ligation-Independent Cloning. Methods Mol. Biol. 2012, 852, 51–59.
- 58 Li, M. Z.; Elledge, S. J. Harnessing Homologous Recombination in Vitro to Generate Recombinant DNA via SLIC. Nat. Methods 2007, 4, 251–256.
- 59 Zhang, Y.; Werling, U.; Edelmann, W. SLiCE: A Novel Bacterial Cell Extract-Based DNA Cloning Method. Nucleic Acids Res. 2012, 40, e55.
- 60 Torella, J. P.; Boehm, C. R.; Lienert, F.; Chen, J. H.; Way, J. C.; Silver, P. A. Rapid Construction of Insulated Genetic Circuits via Synthetic Sequence-Guided Isothermal Assembly. Nucleic Acids Res. 2014, 42, 681–689.
- 61 Torella, J. P.; Lienert, F.; Boehm, C. R.; Chen, J. H.; Way, J. C.; Silver, P. A. Unique Nucleotide Sequence-Guided Assembly of Repetitive DNA Parts for Synthetic Biology Applications. Nat. Protoc. 2014, 9, 2075–2089.
- 62 Trubitsyna, M.; Michlewski, G.; Cai, Y.; Elfick, A.; French, C. E. PaperClip: Rapid Multi-Part DNA Assembly from Existing Libraries. Nucleic Acids Res. 2014, 42, e154.
- 63 Kouprina, N.; Larionov, V. Innovation - TAR Cloning: Insights into Gene Function, Long-Range Haplotypes and Genome Structure and Evolution. Nat. Rev. Genet. 2006, 7, 805–812.
- 64 Gust, B.; Challis, G. L.; Fowler, K.; Kieser, T.; Chater, K. F. PCR-Targeted Streptomyces Gene Replacement Identifies a Protein Domain Needed for Biosynthesis of the Sesquiterpene Soil Odor Geosmin. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 1541–1546.
- 65 Fu, J.; Bian, X.; Hu, S.; Wang, H.; Huang, F.; Seibert, P. M.; Plaza, A.; Xia, L.; Muller, R.; Stewart, A. F.; Zhang, Y. Full-Length RecE Enhances Linear-Linear Homologous Recombination and Facilitates Direct Cloning for Bioprospecting. Nat. Biotechnol. 2012, 30, 440–446.
- 66 Gibson, D. G.; Benders, G. A.; Andrews-Pfannkoch, C.; Denisova, E. A.; Baden-Tillson, H.; Zaveri, J.; Stockwell, T. B.; Brownley, A.; Thomas, D. W.; Algire, M. A.; Merryman, C.; Young, L.; Noskov, V. N.; Glass, J. I.; Venter, J. C.; Hutchison, C. A., 3rd; Smith, H. O. Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma Genitalium Genome. Science 2008, 319, 1215–1220.
- 67 Annaluru, N.; Muller, H.; Mitchell, L. A.; Ramalingam, S.; Stracquadanio, G.; Richardson, S. M.; Dymond, J. S.; Kuang, Z.; Scheifele, L. Z.; Cooper, E. M.; Cai, Y.; Zeller, K.; Agmon, N.; Han, J. S.; Hadjithomas, M.; Tullman, J.; Caravelli, K.; Cirelli, K.; Guo, Z.; London, V.; Yeluru, A.; Murugan, S.; Kandavelou, K.; Agier, N.; Fischer, G.; Yang, K.; Martin, J. A.; Bilgel, M.; Bohutski, P.; Boulier, K. M.; Capaldo, B. J.; Chang, J.; Charoen, K.; Choi, W. J.; Deng, P.; DiCarlo, J. E.; Doong, J.; Dunn, J.; Feinberg, J. I.; Fernandez, C.; Floria, C. E.; Gladowski, D.; Hadidi, P.; Ishizuka, I.; Jabbari, J.; Lau, C. Y.; Lee, P. A.; Li, S.; Lin, D.; Linder, M. E.; Ling, J.; Liu, J.; Liu, J.; London, M.; Ma, H.; Mao, J.; McDade, J. E.; McMillan, A.; Moore, A. M.; Oh, W. C.; Ouyang, Y.; Patel, R.; Paul, M.; Paulsen, L. C.; Qiu, J.; Rhee, A.; Rubashkin, M. G.; Soh, I. Y.; Sotuyo, N. E.; Srinivas, V.; Suarez, A.; Wong, A.; Wong, R.; Xie, W. R.; Xu, Y.; Yu, A. T.; Koszul, R.; Bader, J. S.; Boeke, J. D.; Chandrasegaran, S. Total Synthesis of a Functional Designer Eukaryotic Chromosome. Science 2014, 344, 55–58.
- 68 Richardson, S. M.; Mitchell, L. A.; Stracquadanio, G.; Yang, K.; Dymond, J. S.; DiCarlo, J. E.; Lee, D.; Huang, C. L.; Chandrasegaran, S.; Cai, Y.; Boeke, J. D.; Bader, J. S. Design of a Synthetic Yeast Genome. Science 2017, 355, 1040–1044.
- 69 Ma, Y.; Su, S.; Fu, Z.; Zhou, C.; Qiao, B.; Wu, Y.; Yuan, Y. J. Convenient Synthesis and Delivery of a Megabase-Scale Designer Accessory Chromosome Empower Biosynthetic Capacity. Cell Res. 2024, 34, 309–322.
- 70 Zhou, J.; Wu, R.; Xue, X.; Qin, Z. CasHRA (Cas9-facilitated Homologous Recombination Assembly) Method of Constructing Megabase-Sized DNA. Nucleic Acids Res. 2016, 44, e124.
- 71 He, B.; Ma, Y.; Tian, F.; Zhao, G. R.; Wu, Y.; Yuan, Y. J. YLC-Assembly: Large DNA Assembly via Yeast Life Cycle. Nucleic Acids Res. 2023, 51, 8283–8292.
- 72 Herskowitz, I. Life Cycle of the Budding Yeast Saccharomyces cerevisiae. Microbiol. Rev. 1988, 52, 536–553.
- 73 Lee, N. C.; Larionov, V.; Kouprina, N. Highly Efficient CRISPR/Cas9-Mediated TAR Cloning of Genes and Chromosomal Loci from Complex Genomes in Yeast. Nucleic Acids Res. 2015, 43, e55.
- 74 Larionov, V.; Kouprina, N.; Eldarov, M.; Perkins, E.; Porter, G.; Resnick, M. A. Transformation-Associated Recombination Between Diverged and Homologous DNA Repeats is Induced by Strand Breaks. Yeast 1994, 10, 93–104.
- 75 Mitchell, L. A.; Wang, A.; Stracquadanio, G.; Kuang, Z.; Wang, X.; Yang, K.; Richardson, S.; Martin, J. A.; Zhao, Y.; Walker, R.; Luo, Y.; Dai, H.; Dong, K.; Tang, Z.; Yang, Y.; Cai, Y.; Heguy, A.; Ueberheide, B.; Fenyo, D.; Dai, J.; Bader, J. S.; Boeke, J. D. Synthesis, Debugging, and Effects of Synthetic Chromosome Consolidation: SynVI and Beyond. Science 2017, 355, eaaf4831.
- 76 Shen, Y.; Wang, Y.; Chen, T.; Gao, F.; Gong, J.; Abramczyk, D.; Walker, R.; Zhao, H.; Chen, S.; Liu, W.; Luo, Y.; Muller, C. A.; Paul-Dubois-Taine, A.; Alver, B.; Stracquadanio, G.; Mitchell, L. A.; Luo, Z.; Fan, Y.; Zhou, B.; Wen, B.; Tan, F.; Wang, Y.; Zi, J.; Xie, Z.; Li, B.; Yang, K.; Richardson, S. M.; Jiang, H.; French, C. E.; Nieduszynski, C. A.; Koszul, R.; Marston, A. L.; Yuan, Y.; Wang, J.; Bader, J. S.; Dai, J.; Boeke, J. D.; Xu, X.; Cai, Y.; Yang, H. Deep Functional Analysis of SynII, a 770-Kilobase Synthetic Yeast Chromosome. Science 2017, 355, eaaf4791.
- 77Xie, Z. X.; Li, B. Z.; Mitchell, L. A.; Wu, Y.; Qi, X.; Jin, Z.; Jia, B.; Wang, X.; Zeng, B. X.; Liu, H. M.; Wu, X. L.; Feng, Q.; Zhang, W. Z.; Liu, W.; Ding, M. Z.; Li, X.; Zhao, G. R.; Qiao, J. J.; Cheng, J. S.; Zhao, M.; Kuang, Z.; Wang, X.; Martin, J. A.; Stracquadanio, G.; Yang, K.; Bai, X.; Zhao, J.; Hu, M. L.; Lin, Q. H.; Zhang, W. Q.; Shen, M. H.; Chen, S.; Su, W.; Wang, E. X.; Guo, R.; Zhai, F.; Guo, X. J.; Du, H. X.; Zhu, J. Q.; Song, T. Q.; Dai, J. J.; Li, F. F.; Jiang, G. Z.; Han, S. L.; Liu, S. Y.; Yu, Z. C.; Yang, X. N.; Chen, K.; Hu, C.; Li, D. S.; Jia, N.; Liu, Y.; Wang, L. T.; Wang, S.; Wei, X. T.; Fu, M. Q.; Qu, L. M.; Xin, S. Y.; Liu, T.; Tian, K. R.; Li, X. N.; Zhang, J. H.; Song, L. X.; Liu, J. G.; Lv, J. F.; Xu, H.; Tao, R.; Wang, Y.; Zhang, T. T.; Deng, Y. X.; Wang, Y. R.; Li, T.; Ye, G. X.; Xu, X. R.; Xia, Z. B.; Zhang, W.; Yang, S. L.; Liu, Y. L.; Ding, W. Q.; Liu, Z. N.; Zhu, J. Q.; Liu, N. Z.; Walker, R.; Luo, Y.; Wang, Y.; Shen, Y.; Yang, H.; Cai, Y.; Ma, P. S.; Zhang, C. T.; Bader, J. S.; Boeke, J. D.; Yuan, Y. J. "Perfect" Designer Chromosome V and Behavior of a Ring Derivative. Science 2017, 355, eaaf4704.
- 78 Wu, Y.; Li, B. Z.; Zhao, M.; Mitchell, L. A.; Xie, Z. X.; Lin, Q. H.; Wang, X.; Xiao, W. H.; Wang, Y.; Zhou, X.; Liu, H.; Li, X.; Ding, M. Z.; Liu, D.; Zhang, L.; Liu, B. L.; Wu, X. L.; Li, F. F.; Dong, X. T.; Jia, B.; Zhang, W. Z.; Jiang, G. Z.; Liu, Y.; Bai, X.; Song, T. Q.; Chen, Y.; Zhou, S. J.; Zhu, R. Y.; Gao, F.; Kuang, Z.; Wang, X.; Shen, M.; Yang, K.; Stracquadanio, G.; Richardson, S. M.; Lin, Y.; Wang, L.; Walker, R.; Luo, Y.; Ma, P. S.; Yang, H.; Cai, Y.; Dai, J.; Bader, J. S.; Boeke, J. D.; Yuan, Y. J. Bug Mapping and Fitness Testing of Chemically Synthesized Chromosome X. Science 2017, 355, eaaf4706.
- 79 Zhang, W.; Zhao, G.; Luo, Z.; Lin, Y.; Wang, L.; Guo, Y.; Wang, A.; Jiang, S.; Jiang, Q.; Gong, J.; Wang, Y.; Hou, S.; Huang, J.; Li, T.; Qin, Y.; Dong, J.; Qin, Q.; Zhang, J.; Zou, X.; He, X.; Zhao, L.; Xiao, Y.; Xu, M.; Cheng, E.; Huang, N.; Zhou, T.; Shen, Y.; Walker, R.; Luo, Y.; Kuang, Z.; Mitchell, L. A.; Yang, K.; Richardson, S. M.; Wu, Y.; Li, B. Z.; Yuan, Y. J.; Yang, H.; Lin, J.; Chen, G. Q.; Wu, Q.; Bader, J. S.; Cai, Y.; Boeke, J. D.; Dai, J. Engineering the Ribosomal DNA in a Megabase Synthetic Chromosome. Science 2017, 355, eaaf3981.
- 80 Herrmann, S.; Siegl, T.; Luzhetska, M.; Petzke, L.; Jilg, C.; Welle, E.; Erb, A.; Leadlay, P. F.; Bechthold, A.; Luzhetskyy, A. Site-Specific Recombination Strategies for Engineering Actinomycete Genomes. Appl. Environ. Microb. 2012, 78, 1804–1812.
- 81 Colloms, S. D.; Merrick, C. A.; Olorunniji, F. J.; Stark, W. M.; Smith, M. C.; Osbourn, A.; Keasling, J. D.; Rosser, S. J. Rapid Metabolic Pathway Assembly and Modification Using Serine Integrase Site-Specific Recombination. Nucleic Acids Res. 2014, 42, e23.
- 82 Nour-Eldin, H. H.; Geu-Flores, F.; Halkier, B. A. USER Cloning and USER Fusion: the Ideal Cloning Techniques for Small and Big Laboratories. Methods Mol. Biol. 2010, 643, 185–200.
- 83 de Kok, S.; Stanton, L. H.; Slaby, T.; Durot, M.; Holmes, V. F.; Patel, K. G.; Platt, D.; Shapland, E. B.; Serber, Z.; Dean, J.; Newman, J. D.; Chandran, S. S. Rapid and Reliable DNA Assembly via Ligase Cycling Reaction. ACS Synth. Biol. 2014, 3, 97–106.
- 84 Thieme, F.; Engler, C.; Kandzia, R.; Marillonnet, S. Quick and Clean Cloning: a Ligation-Independent Cloning Strategy for Selective Cloning of Specific PCR Products from Non-Specific Mixes. PLoS One 2011, 6, e20556.
- 85 Bond, S. R.; Naus, C. C. RF-Cloning.org: an OnlineTool for The Design of Restriction-Free Cloning Projects. Nucleic Acids Res. 2012, 40, W209-W213.
- 86 Erijman, A.; Dantes, A.; Bernheim, R.; Shifman, J. M.; Peleg, Y. Transfer-PCR (TPCR): a Highway for DNA Cloning and Protein Engineering. J. Struct. Biol. 2011, 175, 171–177.
- 87 Hillson, N. J.; Rosengarten, R. D.; Keasling, J. D. j5 DNA Assembly Design Automation Software. ACS Synth. Biol. 2012, 1, 14–21.




