Consecutive Methylation Catalyzed by TsrM, an Atypical Class B Radical SAM Methylase
Runze Wu
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorCorresponding Author
Wei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]; [email protected]Search for more papers by this authorRunze Wu
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorCorresponding Author
Wei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]; [email protected]Search for more papers by this authorComprehensive Summary
TsrM is a cobalamin-dependent radical S-adenosylmethionine (SAM) methyltransferase belonging to the Class B radical SAM methylase (RSM) family. This enzyme catalyzes the C-2 methylation of L-tryptophan to produce 2-methyltrytophan (2-MeTrp), an intermediate involved in the biosynthesis of thiostrepton A. In this work, we report characterization of an unexpected activity of TsrM, which carries out an additional methylation reaction on the product 2-MeTrp. A series of isotopic labeling studies and assays with different Trp analogs revealed that TsrM is able to transfer a methyl group from SAM to the C4 of 2-MeTrp to produce 2,4-dimethyltryptophan. These results reveal the intriguing substrate specificity of TsrM, further expanding the reaction promiscuity of the radical SAM superfamily enzymes.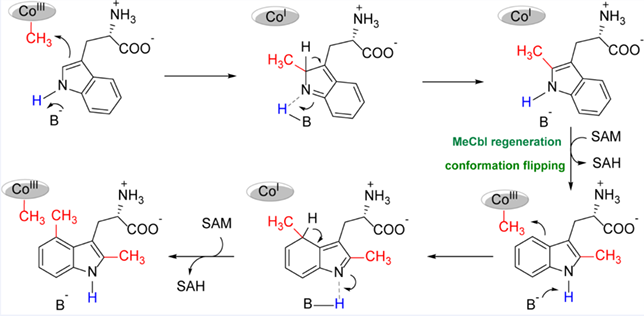
Supporting Information
| Filename | Description |
|---|---|
| cjoc202200174-sup-0001-Supinfo.pdfPDF document, 913.8 KB |
Appendix S1 Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1 Gerlt, J. A.; Bouvier, J. T.; Davidson, D. B.; Imker, H. J.; Sadkhin, B.; Slater, D. R.; Whalen, K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037.
- 2 Frey, P. A.; Hegeman, A. D.; Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 63–88.
- 3 Broderick, J. B.; Duffus, B. R.; Duschene, K. S.; Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317.
- 4 Mahanta, N.; Fedoseyenko, D.; Dairi, T.; Begley, T. P. Menaquinone biosynthesis: formation of aminofutalosine requires a unique radical SAM enzyme. J. Am. Chem. Soc. 2013, 135, 15318–15321.
- 5 Sato, S.; Kudo, F.; Rohmer, M.; Eguchi, T. Characterization of Radical SAM Adenosylhopane Synthase, HpnH, which Catalyzes the 5'-Deoxyadenosyl Radical Addition to Diploptene in the Biosynthesis of C35 Bacteriohopanepolyols. Angew. Chem. Int. Ed. 2020, 59, 237–241.
- 6 Zhong, Y.; Ji, X.; Zhang, Q. Radical SAM-dependent adenosylation involved in bacteriohopanepolyol biosynthesis. Chin. J. Chem. 2020, 38, 39–42.
- 7 Ding, W.; Ji, X.; Zhong, Y.; Xu, K.; Zhang, Q. Adenosylation reactions catalyzed by the radical S-adenosylmethionine superfamily enzymes. Curr. Opin. Chem. Biol. 2020, 55, 86–95.
- 8 Lundahl, M. N.; Sarksian, R.; Yang, H.; Jodts, R. J.; Pagnier, A.; Smith, D. F.; Mosquera, M. A.; van der Donk, W. A.; Hoffman, B. M.; Broderick, W. E.; Broderick, J. B. Mechanism of Radical S-Adenosyl-l-methionine Adenosylation: Radical Intermediates and the Catalytic Competence of the 5'-Deoxyadenosyl Radical. J. Am. Chem. Soc. 2022, 144, 5087–5098.
- 9 Rohac, R.; Amara, P.; Benjdia, A.; Martin, L.; Ruffie, P.; Favier, A.; Berteau, O.; Mouesca, J. M.; Fontecilla-Camps, J. C.; Nicolet, Y. Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE. Nat. Chem. 2016, 8, 491–500.
- 10 Ji, W.; Ji, X.; Zhang, Q.; Mandalapu, D.; Deng, Z.; Ding, W.; Sun, P.; Zhang, Q. Sulfonium-based Homolytic Substitution Observed for the Radical SAM Enzyme HemN. Angew. Chem. Int. Ed. 2020, 59, 8880–8884.
- 11 Cheng, J.; Ji, W.; Ma, S.; Ji, X.; Deng, Z.; Ding, W.; Zhang, Q. Characterization and Mechanistic Study of the Radical SAM Enzyme ArsS Involved in Arsenosugar Biosynthesis. Angew. Chem. Int. Ed. 2021, 60, 7570–7575.
- 12 Tao, L.; Pattenaude, S. A.; Joshi, S.; Begley, T. P.; Rauchfuss, T. B.; Britt, R. D. Radical SAM Enzyme HydE Generates Adenosylated Fe(I) Intermediates En Route to the [FeFe]-Hydrogenase Catalytic H-Cluster. J. Am. Chem. Soc. 2020, 142, 10841–10848.
- 13 Zhao, J.; Ji, W.; Ji, X.; Zhang, Q. Biochemical Characterization of an Arginine 2,3-Aminomutase with Dual Substrate Specificity. Chin. J. Chem. 2020, 38, 959–962.
- 14 Yin, Y.; Ji, X.; Zhang, Q. The Promiscuous Activity of the Radical SAM Enzyme NosL toward Two Unnatural Substrates. Chin. J. Chem. 2021, 39, 2417–2421.
- 15 Honarmand Ebrahimi, K.; Rowbotham, J. S.; McCullagh, J.; James, W. S. Mechanism of Diol Dehydration by a Promiscuous Radical-SAM Enzyme Homologue of the Antiviral Enzyme Viperin (RSAD2). ChemBioChem 2020, 21, 1605–1612.
- 16 Himes, P. M.; Allen, S. E.; Hwang, S.; Bowers, A. A. Production of Sactipeptides in Escherichia coli: Probing the Substrate Promiscuity of Subtilosin A Biosynthesis. ACS Chem. Biol. 2016, 11, 1737–1744.
- 17 Schramma, K. R.; Seyedsayamdost, M. R. Lysine-Tryptophan Crosslinked Peptides Produced by Radical SAM Enzymes in Pathogenic Streptococci. ACS Chem. Biol. 2017, 12, 922–927.
- 18 Ruszczycky, M. W.; Zhong, A.; Liu, H. W. Following the electrons: peculiarities in the catalytic cycles of radical SAM enzymes. Nat. Prod. Rep. 2018, 35, 615–621.
- 19 Cheng, J.; Ding, W.; Zhang, Q. Radical SAM-dependent Demetallation of Heme. Chin. J. Chem. 2022, 40, 1053–1058.
- 20 Bhandari, D. M.; Fedoseyenko, D.; Begley, T. P. Mechanistic Studies on the Radical SAM Enzyme Tryptophan Lyase (NosL). Methods Enzymol. 2018, 606, 155–178.
- 21 Ding, W.; Ji, X.; Li, Y.; Zhang, Q. Catalytic promiscuity of the radical S-adenosyl-L-methionine enzyme NosL. Front. Chem. 2016, 4, 27.
- 22 Ma, S.; Chen, H.; Li, H.; Ji, X.; Deng, Z.; Ding, W.; Zhang, Q. Post-Translational Formation of Aminomalonate by a Promiscuous Peptide-Modifying Radical SAM Enzyme. Angew. Chem. Int. Ed. 2021, 60, 19957–19964.
- 23 Li, H.; Zhao, J.; Ding, W.; Zhang, Q. Glucuronyl C4 dehydrogenation by the radical SAM enzyme BlsE involved in blasticidin S biosynthesis. Chem. Commun. 2022, 58, 3561–3564.
- 24 Lee, Y. H.; Hou, X.; Chen, R.; Feng, J.; Liu, X.; Ruszczycky, M. W.; Gao, J. M.; Wang, B.; Zhou, J.; Liu, H. W. Radical S-Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S. J. Am. Chem. Soc. 2022, 144, 4478–4486.
- 25
Bandarian, V. Journey on the Radical SAM Road as an Accidental Pilgrim. ACS Bio Med Chem Au 2022, doi: https://doi.org/10.1021/acsbiomedchemau.1c00059.
10.1021/acsbiomedchemau.1c00059 Google Scholar
- 26 Zhang, Q.; van der Donk, W. A.; Liu, W. Radical-Mediated Enzymatic Methylation: A Tale of Two SAMS. Acc. Chem. Res. 2012, 45, 555–564.
- 27 Bauerle, M. R.; Schwalm, E. L.; Booker, S. J. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J. Biol. Chem. 2015, 290, 3995–4002.
- 28 Yan, F.; Fujimori, D. G. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3930–3934.
- 29 Grove, T. L.; Benner, J. S.; Radle, M. I.; Ahlum, J. H.; Landgraf, B. J.; Krebs, C.; Booker, S. J. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 2011, 332, 604–607.
- 30 Stojkovic, V.; Fujimori, D. G. Radical SAM-Mediated Methylation of Ribosomal RNA. Methods Enzymol. 2015, 560, 355–376.
- 31 Zhou, S.; Alkhalaf, L. M.; de Los Santos, E. L.; Challis, G. L. Mechanistic insights into class B radical-S-adenosylmethionine methylases: ubiquitous tailoring enzymes in natural product biosynthesis. Curr. Opin. Chem. Biol. 2016, 35, 73–79.
- 32 Ding, W.; Li, Q.; Jia, Y.; Ji, X.; Qianzhu, H.; Zhang, Q. Emerging Diversity of the Cobalamin-Dependent Methyltransferases Involving Radical-Based Mechanisms. ChemBioChem 2016, 17, 1191–1197.
- 33 Sinner, E. K.; Marous, D. R.; Townsend, C. A. Evolution of Methods for the Study of Cobalamin-Dependent Radical SAM Enzymes. ACS Bio Med Chem Au 2022, 2, 4–10.
- 34
Bridwell-Rabb, J.; Li, B.; Drennan, C. L. Cobalamin-Dependent Radical S-Adenosylmethionine Enzymes: Capitalizing on Old Motifs for New Functions. ACS Bio Med Chem Au 2022, doi:https://doi.org/10.1021/acsbiomedchemau.1c00051.
10.1021/acsbiomedchemau.1c00051 Google Scholar
- 35 Jin, W. B.; Wu, S.; Xu, Y. F.; Yuan, H.; Tang, G. L. Recent advances in HemN-like radical S-adenosyl-l-methionine enzyme-catalyzed reactions. Nat. Prod. Rep. 2019, 37, 17–28.
- 36
Cheng, J.; Liu, W.-Q.; Zhu, X.; Zhang, Q. Functional Diversity of HemN-like Proteins. ACS Bio Med Chem Au 2022, doi: 10.1021/ acsbiomedchemau.1c00058.
10.1021/acsbiomedchemau.1c00058 Google Scholar
- 37
Mathew, L. G.; Brimberry, M.; Lanzilotta, W. N. Class C Radical SAM Methyltransferases Involved in Anaerobic Heme Degradation. ACS Bio Med Chem Au 2022, doi: https://doi.org/10.1021/acsbiomedchemau.1c00047.
10.1021/acsbiomedchemau.1c00047 Google Scholar
- 38 Ma, S.; Mandalapu, D.; Wang, S.; Zhang, Q. Biosynthesis of cyclopropane in natural products. Nat. Prod. Rep. 2021, doi: 10.1039/ d1np00065a.
- 39 Allen, K. D.; Xu, H.; White, R. H. Identification of a unique radical S-adenosylmethionine methylase likely involved in methanopterin biosynthesis in Methanocaldococcus jannaschii. J. Bacteriol. 2014, 196, 3315–3323.
- 40 Hu, Y.; Ribbe, M. W. Maturation of nitrogenase cofactor-the role of a class E radical SAM methyltransferase NifB. Curr. Opin. Chem. Biol. 2016, 31, 188–194.
- 41 Pierre, S.; Guillot, A.; Benjdia, A.; Sandstrom, C.; Langella, P.; Berteau, O. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol. 2012, 8, 957–959.
- 42 Benjdia, A.; Pierre, S.; Gherasim, C.; Guillot, A.; Carmona, M.; Amara, P.; Banerjee, R.; Berteau, O. The thiostrepton A tryptophan methyltransferase TsrM catalyses a cob(II)alamin-dependent methyl transfer reaction. Nat. Commun. 2015, 6, 8377.
- 43 Blaszczyk, A. J.; Silakov, A.; Zhang, B.; Maiocco, S. J.; Lanz, N. D.; Kelly, W. L.; Elliott, S. J.; Krebs, C.; Booker, S. J. Spectroscopic and Electrochemical Characterization of the Iron-Sulfur and Cobalamin Cofactors of TsrM, an Unusual Radical S-Adenosylmethionine Methylase. J. Am. Chem. Soc. 2016, 138, 3416–3426.
- 44 Blaszczyk, A. J.; Wang, B.; Silakov, A.; Ho, J. V.; Booker, S. J. Efficient methylation of C2 in l-tryptophan by the cobalamin-dependent radical S-adenosylmethionine methylase TsrM requires an unmodified N1 amine. J. Biol. Chem. 2017, 292, 15456–15467.
- 45 Blaszczyk, A. J.; Knox, H. L.; Booker, S. J. Understanding the role of electron donors in the reaction catalyzed by Tsrm, a cobalamin-dependent radical S-adenosylmethionine methylase. J. Biol. Inorg. Chem. 2019, 24, 831–839.
- 46 Ulrich, E. C.; Drennan, C. L. The Atypical Cobalamin-Dependent S-Adenosyl-l-Methionine Nonradical Methylase TsrM and Its Radical Counterparts. J. Am. Chem. Soc. 2022, 144, 5673–5684.
- 47 Liao, R.; Duan, L.; Lei, C.; Pan, H.; Ding, Y.; Zhang, Q.; Chen, D.; Shen, B.; Yu, Y.; Liu, W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009, 16, 141–147.
- 48 Kelly, W. L.; Pan, L.; Li, C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 2009, 131, 4327–4334.
- 49 Duan, L.; Wang, S.; Liao, R.; Liu, W. Insights into quinaldic acid moiety formation in thiostrepton biosynthesis facilitating fluorinated thiopeptide generation. Chem. Biol. 2012, 19, 443–448.
- 50 Montalban-Lopez, M.; Scott, T. A.; Ramesh, S.; Rahman, I. R.; van Heel, A. J.; Viel, J. H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; Grande Burgos, M. J.; Hill, C.; Kim, S.; Koehnke, J.; Latham, J. A.; Link, A. J.; Martinez, B.; Nair, S. K.; Nicolet, Y.; Rebuffat, S.; Sahl, H. G.; Sareen, D.; Schmidt, E. W.; Schmitt, L.; Severinov, K.; Sussmuth, R. D.; Truman, A. W.; Wang, H.; Weng, J. K.; van Wezel, G. P.; Zhang, Q.; Zhong, J.; Piel, J.; Mitchell, D. A.; Kuipers, O. P.; van der Donk, W. A. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239.
- 51 Knox, H. L.; Chen, P. Y.; Blaszczyk, A. J.; Mukherjee, A.; Grove, T. L.; Schwalm, E. L.; Wang, B.; Drennan, C. L.; Booker, S. J. Structural basis for non-radical catalysis by TsrM, a radical SAM methylase. Nat. Chem. Biol. 2021, 17, 485-491.
- 52 Dong, Y.; Zhang, S.; Zhao, L. Unravelling the Structural Development of Peptide-Coordinated Iron-Sulfur Clusters: Prebiotic Evolution and Biosynthetic Strategies. Chin. J. Chem. 2022, 40, 1478–1491.
- 53 Buller, A. R.; Brinkmann-Chen, S.; Romney, D. K.; Herger, M.; Murciano-Calles, J.; Arnold, F. H. Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation. Proc. Natl. Acad. Sci. 2015, 112, 14599–14604.
- 54 Parent, A.; Guillot, A.; Benjdia, A.; Chartier, G.; Leprince, J.; Berteau, O. The B12-Radical SAM Enzyme PoyC Catalyzes Valine Cbeta-Methylation during Polytheonamide Biosynthesis. J. Am. Chem. Soc. 2016, 138, 15515–15518.
- 55 Wang, Y.; Schnell, B.; Baumann, S.; Muller, R.; Begley, T. P. Biosynthesis of Branched Alkoxy Groups: Iterative Methyl Group Alkylation by a Cobalamin-Dependent Radical SAM Enzyme. J. Am. Chem. Soc. 2017, 139, 1742–1745.
- 56 Wang, Y.; Begley, T. P. Mechanistic Studies on CysS - A Vitamin B12-Dependent Radical SAM Methyltransferase Involved in the Biosynthesis of the tert-Butyl Group of Cystobactamid. J. Am. Chem. Soc. 2020, 142, 9944–9954.
- 57 Marous, D. R.; Lloyd, E. P.; Buller, A. R.; Moshos, K. A.; Grove, T. L.; Blaszczyk, A. J.; Booker, S. J.; Townsend, C. A. Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis. Proc. Natl. Acad. Sci. 2015, 112, 10354–10358.
- 58 Sinner, E. K.; Lichstrahl, M. S.; Li, R.; Marous, D. R.; Townsend, C. A. Methylations in complex carbapenem biosynthesis are catalyzed by a single cobalamin-dependent radical S-adenosylmethionine enzyme. Chem. Commun. 2019, 55, 14934–14937.
- 59 Knox, H. L.; Sinner, E. K.; Townsend, C. A.; Boal, A. K.; Booker, S. J. Structure of a B12-dependent radical SAM enzyme in carbapenem biosynthesis. Nature 2022, 602, 343–348.
- 60 Ji, X.; Mo, T.; Liu, W. Q.; Ding, W.; Deng, Z.; Zhang, Q. Revisiting the Mechanism of the Anaerobic Coproporphyrinogen III Oxidase HemN. Angew. Chem. Int. Ed. 2019, 58, 6235–6238.
- 61 Soualmia, F.; Guillot, A.; Sabat, N.; Brewee, C.; Kubiak, X.; Haumann, M.; Guinchard, X.; Benjdia, A.; Berteau, O. Exploring the biosynthetic capacity of TsrM, a B12-dependent Radical SAM Enzyme Catalyzing Non-radical Reactions. Chem. - Eur. J. 2022, doi: https://doi.org/10.1002/chem.202200627.
- 62 McLaughlin, M. I.; van der Donk, W. A. Stereospecific Radical-Mediated B12-Dependent Methyl Transfer by the Fosfomycin Biosynthesis Enzyme Fom3. Biochemistry 2018, 57, 4967–4971.
- 63 Blaszczyk, A. J.; Wang, R. X.; Booker, S. J. TsrM as a Model for Purifying and Characterizing Cobalamin-Dependent Radical S-Adenosylmethionine Methylases. Methods Enzymol. 2017, 595, 303–329.




