Supramolecular Genome Editing: Targeted Delivery and Endogenous Activation of CRISPR/Cas9 by Dynamic Host-Guest Recognition
Bowen Li
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
The authors contributed equally to this work.
Contribution: Conceptualization (lead), Data curation (lead), Writing - original draft (lead)
Search for more papers by this authorQing Li
Stoddart Institute of Molecular Science, Department of Chemistry, Zhejiang University, Hangzhou, 310058 P. R. China
Zhejiang-Israel Joint Laboratory of Self-Assembling Functional Materials, ZJU-Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, 311215 P. R. China
The authors contributed equally to this work.
Contribution: Conceptualization (equal), Formal analysis (lead), Software (lead), Writing - original draft (supporting)
Search for more papers by this authorZidan Qi
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Investigation (supporting), Methodology (supporting)
Search for more papers by this authorZhiyao Li
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Formal analysis (supporting), Methodology (equal), Resources (lead)
Search for more papers by this authorXiaojie Yan
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Conceptualization (supporting), Investigation (lead), Validation (equal)
Search for more papers by this authorYuan Chen
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Formal analysis (equal), Methodology (supporting), Visualization (lead)
Search for more papers by this authorXiaojie Xu
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
Contribution: Methodology (equal), Validation (supporting), Visualization (supporting)
Search for more papers by this authorQi Pan
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Data curation (supporting), Methodology (supporting), Resources (supporting)
Search for more papers by this authorYuxuan Chen
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Investigation (supporting), Methodology (supporting), Software (equal)
Search for more papers by this authorCorresponding Author
Feihe Huang
Stoddart Institute of Molecular Science, Department of Chemistry, Zhejiang University, Hangzhou, 310058 P. R. China
Zhejiang-Israel Joint Laboratory of Self-Assembling Functional Materials, ZJU-Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, 311215 P. R. China
Contribution: Investigation (equal), Project administration (equal), Supervision (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Yuan Ping
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
Contribution: Investigation (equal), Project administration (lead), Supervision (lead), Writing - original draft (equal), Writing - review & editing (lead)
Search for more papers by this authorBowen Li
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
The authors contributed equally to this work.
Contribution: Conceptualization (lead), Data curation (lead), Writing - original draft (lead)
Search for more papers by this authorQing Li
Stoddart Institute of Molecular Science, Department of Chemistry, Zhejiang University, Hangzhou, 310058 P. R. China
Zhejiang-Israel Joint Laboratory of Self-Assembling Functional Materials, ZJU-Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, 311215 P. R. China
The authors contributed equally to this work.
Contribution: Conceptualization (equal), Formal analysis (lead), Software (lead), Writing - original draft (supporting)
Search for more papers by this authorZidan Qi
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Investigation (supporting), Methodology (supporting)
Search for more papers by this authorZhiyao Li
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Formal analysis (supporting), Methodology (equal), Resources (lead)
Search for more papers by this authorXiaojie Yan
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Conceptualization (supporting), Investigation (lead), Validation (equal)
Search for more papers by this authorYuan Chen
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Formal analysis (equal), Methodology (supporting), Visualization (lead)
Search for more papers by this authorXiaojie Xu
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
Contribution: Methodology (equal), Validation (supporting), Visualization (supporting)
Search for more papers by this authorQi Pan
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Data curation (supporting), Methodology (supporting), Resources (supporting)
Search for more papers by this authorYuxuan Chen
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Contribution: Investigation (supporting), Methodology (supporting), Software (equal)
Search for more papers by this authorCorresponding Author
Feihe Huang
Stoddart Institute of Molecular Science, Department of Chemistry, Zhejiang University, Hangzhou, 310058 P. R. China
Zhejiang-Israel Joint Laboratory of Self-Assembling Functional Materials, ZJU-Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, 311215 P. R. China
Contribution: Investigation (equal), Project administration (equal), Supervision (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Yuan Ping
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058 P. R. China
Liangzhu Laboratory, Zhejiang University, Hangzhou, 311121 P. R. China
Contribution: Investigation (equal), Project administration (lead), Supervision (lead), Writing - original draft (equal), Writing - review & editing (lead)
Search for more papers by this authorGraphical Abstract
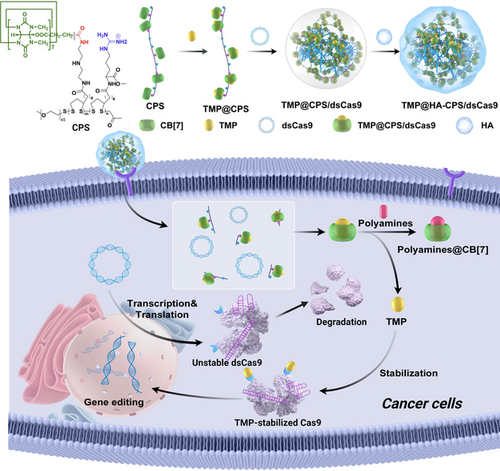
We synthesized a kind of supramolecular cationic polymer bearing cucurbit[7]uril (CB[7]) for the delivery of the plasmid encoding destabilized Cas9 (dsCas9) and single-guide RNA. The dynamic host-guest complexation between CB[7] moieties and trimethoprim, a stabilizer for dsCas9, can be competitively decomplexed by elevated polyamines in the tumor cells to activate Cas9-based genome editing in vivo, thereby ensuring the highly specific therapeutic genome editing for cancer gene therapy.
Abstract
We synthesize supramolecular poly(disulfide) (CPS) containing covalently attached cucurbit[7]uril (CB[7]), which is exploited not only as a carrier to deliver plasmid DNA encoding destabilized Cas9 (dsCas9), but also as a host to include trimethoprim (TMP) by CB[7] moieties through the supramolecular complexation to form TMP@CPS/dsCas9. Once the plasmid is transfected into tumor cells by CPS, the presence of polyamines can competitively trigger the decomplexation of TMP@CPS, thereby displacing and releasing TMP from CB[7] to stabilize dsCas9 that can target and edit the genomic locus of PLK1 to inhibit the growth of tumor cells. Following the systemic administration of TMP@CPS/dsCas9 decorated with hyaluronic acid (HA), tumor-specific editing of PLK1 is detected due to the elevated polyamines in tumor microenvironment, greatly minimizing off-target editing in healthy tissues and non-targeted organs. As the metabolism of polyamines is dysregulated in a wide range of disorders, this study offers a supramolecular approach to precisely control CRISPR/Cas9 functions under particular pathological contexts.
Conflict of interests
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The raw/processed data required to reproduce these findings are available from the authors.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202316323-sup-0001-misc_information.pdf3 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1T. Wan, Y. Ping, Adv. Drug Delivery Rev. 2021, 168, 196–216.
- 2J. A. Doudna, Nature 2020, 578, 229–236.
- 3Y. X. Chen, Y. Ping, Acc. Chem. Res. 2023, 56, 2185–2196.
- 4D. P. Dever, R. O. Bak, A. Reinisch, J. Camarena, G. Washington, C. E. Nicolas, M. Pavel-Dinu, N. Saxena, A. B. Wilkens, S. Mantri, N. Uchida, A. Hendel, A. Narla, R. Majeti, K. I. Weinberg, M. H. Porteus, Nature 2016, 539, 384–389.
- 5T. Carvalho, Nat. Med. 2022, 28, 2438.
- 6J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O'Connell, K. R. Walsh, K. Wood, J. Phillips, Y. X. Xu, A. Amaral, A. P. Boyd, J. E. Cehelsky, M. D. McKee, A. Schiermeier, O. Harari, B. Chir, A. Murphy, C. A. Kyratsous, B. Zambrowicz, R. Soltys, D. E. Gutstein, J. Leonard, L. Sepp-Lorenzino, D. Lebwohl, N. Engl. J. Med. 2021, 385, 493–503.
- 7D. Y. Richards, S. R. Winn, S. Dudley, S. Nygaard, T. L. Mighell, M. Grompe, C. O. Harding, Mol. Ther. Methods Clin. Dev. 2020, 17, 234–245.
- 8J. J. Guo, T. Wan, B. W. Li, Q. Pan, H. H. Xin, Y. Y. Qiu, Y. Ping, ACS Cent. Sci. 2021, 7, 990–1000.
- 9T. Wei, Q. Cheng, Y. L. Min, E. N. Olson, D. J. Sigwart, Nat. Commun. 2020, 11, 3232.
- 10L. Li, S. Hu, X. Y. Chen, Biomaterials 2018, 171, 207–218.
- 11M. Behra, J. Zhou, B. Xu, H. W. Zhang, Acta Pharm. Sin. B 2021, 11, 2150–2171.
- 12W. L. Chew, M. Tabebordbar, J. K. W. Cheng, P. Mali, E. Y. Wu, A. H. M. Ng, K. Zhu, A. J. Wagers, G. M. Church, Nat. Methods 2016, 13, 868–874.
- 13H. Manghwar, B. Li, X. Ding, A. Hussain, K. Lindsey, X. L. Zhang, S. X. Jin, Adv. Sci. 2020, 7, 1902312.
- 14M. Naeem, S. Majeed, M. Z. Hoque, I. Ahmad, Cells 2020, 9, 1608.
- 15L. R. Polstein, C. A. Gersbach, Nat. Chem. Biol. 2015, 11, 198–200.
- 16H. L. Tang, X. J. Xu, Y. X. Chen, H. H. Xin, T. Wan, B. W. Li, H. M. Pan, D. Li, Y. Ping, Adv. Mater. 2021, 33, 2006003.
- 17X. H. Chen, Y. X. Chen, H. H. Xin, T. Wan, Y. Ping, Proc. Nat. Acad. Sci. 2020, 117, 2395–2405.
- 18Y. X. Chen, X. H. Chen, D. Wu, H. H. Xin, D. S. Chen, D. Li, H. M. Pan, C. X. Zhou, Y. Ping, Chem. Mater. 2021, 33, 81–91.
- 19X. J. Yan, Q. Pan, H. H. Xin, Y. X. Chen, Y. Ping, Sci. Adv. 2021, 7, eabj0624.
- 20Y. Yu, X. Wu, N. Guan, J. Shao, H. Li, Y. Chen, Y. Ping, D. Li, H. Ye, Sci. Adv. 2020, 6, eabb1777.
- 21J. Shao, M. Wang, G. Yu, S. Zhu, Y. Yu, B. C. Heng, J. Wu, H. Ye, Proc. Natl. Acad. Sci. USA 2018, 115, E6722–E6730.
- 22Y. Pan, J. Yang, X. Luan, X. Liu, X. Li, J. Yang, T. Huang, L. Sun, Y. Wang, Y. Lin, Y. N. Song, Sci. Adv. 2019, 5, eaav7199.
- 23B. Zetsche, S. E. Volz, F. Zhang, Nat. Biotechnol. 2015, 33, 139–142.
- 24D. Manna, B. Maji, S. A. Gangopadhyay, K. J. Cox, Q. X. Zhou, B. K. Law, R. Mazitschek, A. Choudhary, Angew. Chem. Int. Ed. 2019, 58, 6285–6289.
- 25K. M. Davis, V. Pattanayak, D. B. Thompson, J. A. Zuris, D. R. Liu, Nat. Chem. Biol. 2015, 11, 316–318.
- 26B. Maji, C. L. Moore, B. Zetsche, S. E. Volz, F. Zhang, M. D. Shoulders, Nat. Chem. Biol. 2017, 13, 9–11.
- 27E. W. Gerner, F. L. Meyskens, Nat. Rev. Cancer 2004, 4, 781–792.
- 28Y. Y. Chen, Z. H. Huang, J. F. Xu, Z. W. Sun, X. Zhang, ACS Appl. Mater. Interfaces 2016, 8, 22780–22784.
- 29H. Chen, Y. Y. Chen, H. Wu, J. F. Xu, Z. W. Sun, X. Zhang, Biomaterials 2018, 178, 697–705.
- 30R. Wetzel, S. Shivaprasad, A. D. Williams, Biochemistry 2007, 46, 1–10.
- 31V. Battaglia, C. D. Shields, T. M. Stewart, R. A. Casero, Amino Acids 2014, 46, 511–519.
- 32M. Amin, S. Y. Tang, L. Shalamanova, R. L. Taylor, S. Wylie, B. M. Abdullah, K. A. Whitehead, Biomarkers 2021, 26, 77–94.
- 33R. A. Casero, L. J. Marton, Nat. Rev. Drug Discovery 2007, 6, 373–390.
- 34J. J. Li, Y. Meng, X. L. Wu, Y. X. Sun, Cancer Cell Int. 2020, 20, 539.
- 35R. A. Casero Jr, T. M. Stewart, A. E. Pegg, Nat. Rev. Cancer 2018, 18, 681–695.
- 36K. Soda, J. Exp. Clin. Cancer Res. 2011, 30, 95.
- 37X. Xu, H. L. Tang, J. J. Guo, H. H. Xin, Y. Ping, Signal Transduct. Target. Ther. 2022, 7, 269.
- 38D. Mao, Y. J. Liang, Y. M. Liu, X. H. Zhou, J. Q. Ma, B. Jiang, J. Liu, D. Ma, Angew. Chem. Int. Ed. 2017, 56, 12614–12618.
- 39M. M. Ayhan, H. Karoui, M. Hardy, A. Rockenbauer, L. Charles, R. Rosas, K. Udachin, P. Tordo, D. Bardelang, O. Ouari, J. Am. Chem. Soc. 2015, 137, 10238–10245.
- 40Q. Laurent, R. Martinent, B. Lim, A. Pham, T. Kato, J. López-Andarias, N. Sakai, S. Matile, JACS Au 2021, 1, 710–728.
- 41Y. Lee, T. Ishii, H. J. Kim, N. Nishiyama, Y. Hayakawa, K. Itaka, K. Kataoka, Angew. Chem. 2010, 122, 2606–2609.
- 42R. Shrestha, M. Elsabahy, S. Florez-Malaver, S. Samarajeewa, K. L. Wooley, Biomaterials 2012, 33, 8557–8568.
- 43A. Spadea, J. M. R. Rosa, A. Tirella, M. B. Ashford, K. J. Williams, I. J. Stratford, N. Tirelli, M. Mehibel, Mol. Pharmaceutics 2019, 16, 2481–2493.
- 44Y. Sakurai, H. Harashima, Expert Opin. Drug Delivery 2019, 16, 915–936.
- 45X. G. Liu, M. Wu, Q. L. Hu, H. Z. Bai, S. Q. Zhang, Y. Q. Shen, G. P. Tang, Y. Ping, ACS Nano 2016, 10, 11385–11396.
- 46S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak, W. C. W. Chan, Nat. Rev. Mater. 2016, 1, 16014.
- 47T. Wan, Q. Pan, C. Y. Liu, J. J. Guo, B. W. Li, X. J. Yan, Y. Y. Cheng, Y. Ping, Nano Lett. 2021, 21, 9761–9771.
- 48T. Q. Lang, X. Y. Dong, Z. Zheng, Y. R. Liu, G. R. Wang, Q. Yin, Y. P. Li, Sci. Bull. 2019, 64, 91–100.