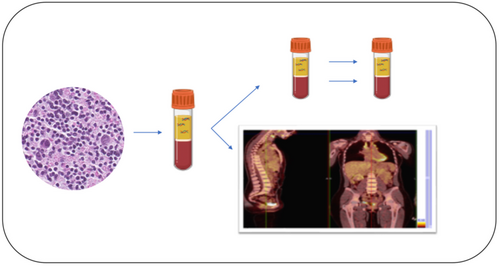
Liquid biopsy: A promising tool for driving strategies and predicting failures in patients with classic Hodgkin lymphoma
Francesco Pepe and Claudia Giordano contributed equally to this work.
Abstract
Classic Hodgkin lymphoma (cHL) consists of a heterogeneous group of haematological disorders that covers undifferentiated B cell neoplasms originating from germinal centre B cells. The HL molecular characterization still represents an ongoing challenge due to the low fraction of tumour Hodgkin and Reed–Sternberg cells mixed with a plethora of non-tumour haematological cells. In this scenario, next generation sequencing of liquid biopsy samples is emerging as a useful tool in HL patients' management. In this review, we aimed to overview the clinical and methodological topics regarding the implementation of molecular analysis in cHL, focusing on the role of liquid biopsy in diagnosis, follow-up, and response prediction.
Graphical Abstract
Liquid biopsy is considered an emerging tool able to integrate conventional tissue specimens for molecular analysis and FDG-PET results to evaluate residual minimal disease and early detection of chemorefractory patients. A combined approach with liquid biopsy may integrate FDG-PET results in order to evaluate residual disease for the early identification of chemorefractory cHL patients.
1 INTRODUCTION
Classic Hodgkin lymphoma (cHL) consists of a heterogeneous group of haematological disorders that covers undifferentiated B cell neoplasms originating from germinal centre B cells.1 To date, the diagnosis of cHL is carried out by inspecting histological and cytological samples but lineage identification represents an ongoing challenge due to low fraction (ranging from 0.1% to 10.0%) of tumour Hodgkin and Reed–Sternberg (HRS) cells mixed with a plethora of non-tumour haematological cells (e.g., eosinophils, macrophages, B and T lymphocytes).1-3 In this scenario, molecular profiling is considered a new weapon aimed at integrating morphological diagnosis. Recently, the activation of JAK/STAT and NF-kB signalling pathways was observed as constitutive mechanisms for cHL progression.4-6 Although formalin-fixed paraffin-embedded (FFPE) samples still remain the most suitable diagnostic material for the implementation of molecular tests, liquid biopsy is considered an emerging tool able to integrate conventional tissue specimens into the clinical setting. Here, we overview clinical and methodological topics related to the implementation of molecular analysis in cHL patients.
2 HL MOLECULAR ANALYSIS: EXPERIENCES WITH FFPE MATERIAL
To date, several assays have been developed to routinely perform molecular analysis in cHL patients. First, Küppers et al7 focused on the evaluation of the mRNA expression profile in preclinical models in order to establish a common signature in cHL patients. This effort was based on the implementation of U95A microarrays (Affymetrix Inc.) on the corresponding platform. Data processing was carried out by using Affymetrix Microarray Suite 4.0 software selecting an arbitrary cutoff of 20 to determine expression level variations.7 Briefly, cell lines from different haematological malignancies highlighted a distinct expression profile able to distinguish cHL from the other groups. Of note, five of the upregulated genes (PRAME, IPL, FER, Rab13, and EAR3) from a previously detected positive group were confirmed in tissue samples from nine cHL patients by adopting a customised real time polymerase chain reaction (RT-PCR) assay.7 Similarly, Malec et al8 investigated the role of inflammation in cHL patients by inspecting pro-inflammatory cytokine levels in 15 patients with heterogeneous morphological features. In this case, an optimised RT-PCR system was enabled to perform molecular analysis. Overall, a matched cytokines expression pattern between preclinical models and patients’ samples was identified. Briefly, IL-10, IL-1b, IL-15 and IL-12p35 were more highly expressed than matched control samples.8 These data demonstrated that cHLs drastically impact the modulation of both the mRNA expression profile of tumour cells and modifications of cytokines levels. These strategies highlighted some significant technical and clinical challenges. First, these technical approaches were able to work successfully only when starting from frozen samples. In addition, the identification of non-recurrent expression signatures indicated that the clinical role of other promising biomarkers should be investigated. In light of these critical issues, the implementation the of next generation sequencing platform (NGS) has rapidly modified the technical paradigm for the molecular characterization of cHL patients (Table 1).7-10 Recently, a whole exome sequencing approach has been validated to comprehensively analyse molecular profiles (DNA and RNA) in a set of 10 cHL patients. In brief, Reichel et al optimised KAPA Biosystems technical workflow integrated with a sorting system in order to selectively improve technical sensitivity in target cells.10 Remarkably, a median number of 244 somatic mutations were detected in each case. Among them, B2M harboured different molecular alterations that inactivated protein function.10 It was observed that B2M loss of function plays a pivotal role as a diagnostic (NS subtype 86/115 cases; 75%) and prognostic biomarker (B2M expression is significantly associated with stage III-IV; p = 0.001) in cHL patients.10 Similarly, Mata et al implemented a deep NGS platform based on a semiconductor system for the molecular analysis of cancer-related genes by adopting a customised panel integrated with a dedicated bioinformatic pipeline.9 Focusing on 57 FFPE samples from cHL patients, CSF2RB, EP300, STAT6, and BTK showed the highest mutation rate according to the most frequent deregulated pathway in cHL patients.9 Moreover, a not negligible percentage of cases (10.3%) was affected by somatic alterations in members of the B cell receptor pathway, considered an emerging target for therapeutic approaches.9 In another experiment, Mata et al11 analysed 35 mostly mutated genes in 20 cHL patients by adopting a semiconductor-based NGS platform for a series of matched original pretreatment and relapse FFPE biopsy samples. Overall, an increasing rate of high impacting p53 molecular alterations and inactivating molecular alterations in epigenetic regulators EP300 and CREBBP were detected in the refractory samples. This study demonstrated the dynamic evolution of the molecular landscape in cHL patients after a conventional treatment strategy.11
| Technique | Assay | Platform | Patients | Sample type | (LoD) % | Setting | References |
|---|---|---|---|---|---|---|---|
| Microarray | GeneChip® Human Genome U95 Set (High Wycombe, United Kingdom) | Affymetrix Inc. (High Wycombe, United Kingdom) | 9 | Frozen tissue | 0.0001 | Discovery (identification of genes involved in the pathogenetic mechanisms of cHL) | Küppers et al7 |
| RT-PCR | Cytokine Gene Expression Plate I (Thermo Fisher Scientifics, USA) | ABI Prism TM 7700 (Thermo Fisher Scientifics, USA) | 16 | Frozen tissue | 0.0001 | Diagnosis (identification of cytokines pattern expressions in cHL patients) | Malec et al8 |
| NGS | WES, KAPA Library Preparation Kits, (Kapa Biosystems, USA) | HiSeq sequencer (Illumina, USA) | 10 | Frozen tissue | NA | Therapy, follow-up (identification of specific genomic alterations for clinical response or to monitor disease clonal evolution) | Reichel et al10 |
| NGS | AmpliSeq Custom Panel (35 genes) | Ion PGM (Thermo Fisher Scientifics, USA) | 57 | FFPE | NA | Therapy, follow-up (identification of new therapeutic targets and molecular patterns in relapsed patients) | Mata et al9 |
| NGS | Custom Panel (35 genes) | Ion PGM (Thermo Fisher Scientifics, USA) | 20 | FFPE, plasma | NA | Follow up (identification of recurrent molecular pathways in refractory cHL patients) | Mata et al11 |
| NGS, CAPP-Seq | Custom Panel (77 genes) | NextSeq500 (Illumina, USA) | 80 untreated plus 32 relapsed/refractory | Plasma, FFPE | 0.0001% | Diagnosis, prognosis and follow-up (Identification of pathogenic alterations, track clonal evolution, MRD | Spina et al20 |
| NGS | Custom Panel (seven genes) | Ion PGM (Thermo Fisher Scientifics, USA) | 60 | Plasma | 1.0% | Follow up (evaluation of MRD in cHL patients) | Camus et al22 |
- Abbreviations: CAPP-Seq, CAncer Personalised Profiling by deep Sequencing; cHL, classical Hodgkin lymphoma; FFPE, formalin-fixed, paraffin-embedded; LoD, limit of detection; MRD, minimal residual disease; NA, not available; NGS, next generation sequencing; RT-PCR, real time polymerase chain reaction; WES, whole exome sequencing.
3 CHL MOLECULAR ANALYSIS: EXPERIENCES ON LIQUID BIOPSY
The liquid biopsy consists of drawn peripheral blood, providing a less invasive, ready-to-use and dynamic sampling modality to investigate a plethora of different biomarkers.12, 13 It was observed that circulating tumour DNA levels (ctDNA) are age- and gender-dependent in lymphoma patients14, 15 with a statistically significant rate higher levels in advanced stage and low responder patients.14, 16 In this regard, NGS platforms include an heterogenous group of sequencing systems that allow simultaneous detection of a broad spectrum of cancer-related genes starting from a low number of nucleic acids fragments.13, 14, 17 Of note, one of the most frequently adopted methods is an ultra-sensitive hybrid selection-based panel (SeqCap EZ Choice Library) on the MiSeq platform (Illumina). This approach is based on selective identification and amplification of exonic and intronic regions that harbour hotspot molecular alterations for a particular cancer type.17-19 Interestingly, Spina et al20 focused on the comprehensive role of liquid biopsy in the diagnosis of cHL without any previous HRS selection, for the identification of the molecular evolution of tumour cells and the detection of minimal residual disease (MRD) after conventional therapeutic strategies. The authors designed and developed a customised NGS panel able to detect hotspot exons and intronic sites in 77 mostly mutated genes in mature B cell tumours on a series of 80 naïve and 32 relapsed or refractory (R/R) first line cHL patients.20 As previously demonstrated, this approach enabled the selective identification of most recurrent cancer-related molecular alterations at very low frequencies.18, 19 Data analysis was optimised according to molecular alteration recurrence. In particular, a mutant allele fraction (MAF) ≥5.0% and ranged between 1.0% and 5.0% for unknown and well-established cancer-related genes were taken in account, respectively.20 Molecular analysis was approached on liquid biopsy samples at different clinically relevant timing points: at baseline, during the course (interim) and at the conclusion of a conventional ABVD (adriamycin, bleomycin, vinblastine, dacarbazine) chemotherapy regimen, or at progression in resistant cHL patients. In addition, genomic DNA extracted from white cells on matched plasma specimens and from neoplastic cells on tissue specimens without any HRS cell selection was collected for molecular testing. Remarkably, TNFAIP3, ITPKB, GNA13, and B2M driver alterations not yet reported in the literature were found in cell free DNA (cfDNA) samples of 15 cHL patients.20 A similar molecular spectrum between newly diagnosed and relapsing cHL patients was observed; this point demonstrated the technical feasibility of this approach for the selective identification of molecular alterations settled in tumour cells. In addition, biopsy specimens also highlighted an agreement of matched results according to cfDNA-based detected alterations (87.5%). On the basis of these results, NGS showed a median of five somatic mutations in 81.2% of newly diagnosed cHL cases; then a median MAF was observed as 5.5% while the vast majority of molecular alterations had a frequency higher than 1.0% (87.3%).20 Moreover, different histological groups matched with specific molecular profiles.
Briefly, a retrospective series of 60 newly diagnosed cHL patients was evaluated by using a semiconductor-based NGS platform that covers hotspot molecular alterations in pivotal genes for haematological malignancies patients.21 Data analysis was carried out by filtering MAF below 0.5%. Overall, a concordance rate of 83.3% (61 variants) was found between tissue and matched cfDNA samples; then, a higher amount of cfDNA was observed in patients with a worse clinical outcome, and in fact, a statistically significant correlation was observed with tumour volume.21
4 LIQUID BIOPSY IN CHL: A PROGNOSTIC TOOL
Currently, there are no biomarkers that are predictive of cHL.22 Standard lymphadenectomy or tissue biopsy is the recommended method for lymphoma diagnosis and is a routine method of oncogene profiling, but the rarity of HRS cells increases the complexity of pathological diagnosis in small biopsy samples. In fact, to date, the genetic landscape of cHL has been only incompletely described because HRS tumour cells are very few.23 Despite the scarcity of HRS cells and a typical lower tumour volume, many studies suggest that cHL seems to display a higher trend of releasing cfDNA than diffuse large B cell lymphoma (DLBCL). This finding may be related to an increased fraction of tumour cells in apoptosis but also related to the nature of the alterations in the nuclear DNA of HRS cells.24
For patients with cHL it is very crucial to discriminate the responders from non-responders at the end of frontline chemotherapy with ABVD, in order to intensify treatment with salvage regimens, and to detect possible relapse in a timely manner.25
Currently, the 2-deoxy-2[F-18]fluoro-D-glucose positron emission tomography (FDG-PET) is considered the standard of care for remission assessment in HL26 and is driving strategies of treatment in patients with cHL. However, the cfDNA baseline assessment might foresee the patient's outcome. Indeed, there is a link between the tumour burden (expressed either as metabolic tumour volume [MTV] or as total metabolic tumour surface or as volume of the bounding box including tumour [TumBB]) and the amount of cfDNA released. Thus, cfDNA could be complementary to FDG-PET staging.27
In 20%-30% of patients interim FDG-PET results do not correspond with the final outcome. In these cases in particular, ct/cfDNA may come to the aid, being potentially complementary or even a convenient substitute for imaging.14, 27
As a matter of fact, in the recent years, studies on liquid biopsy have proven that the peripheral blood level of ctDNA at interim (after two cycles) or at the end of treatment (EOT) strictly correlates with disease response and that it could be a potential tool for evaluating the MRD in the real-world setting. Spina et al20 demonstrated that a 2-log drop in cfDNA between diagnosis and at the end of the second ABVD course can predict metabolic complete response, in particular when the cfDNA trend did not match with interim PET/TC results. The 2-log drop threshold was borrowed from expertise on DLBCL and is named early molecular response—EMR (after 1 cycle of treatment). Another threshold used for DLBCL is a 2.5-log drop after 2 cycles of treatment, named major molecular response (MMR).28
In the same way, in the report by Camus et al a consistent decreasing of cfDNA levels (−78.8%) was identified in 45 patients under chemotherapy regimens (83.0%). Finally, the CAncer Personalised Profiling by deep Sequencing (CAPP-Seq) system was able to distinguish molecular patterns representative of tumour staging and evolution suggesting that ultra-deep NGS systems may represent a solid strategy for investigating liquid biopsy samples in different clinical settings. In light of these results, other studies have demonstrated the role of integrated CAPP-Seq workflow for liquid biopsy analysis in monitoring residual disease setting. These trials focused on a comparison between FDG-PET and molecular analysis on liquid biopsy specimens to select patients that could be treated according to relapsing risk. Interestingly, a positive and negative predictive value of 50.0% and 70.0% was detected in comparison with FDG-PET.21
According to many trials (GHSG HD18 trial, LYSA AHL2011, RATHL trial) interim PET/CT results guide the subsequent treatment, either in terms of intensification to improve the response (GHSG HD18 trial), or in terms of de-escalation when a response is already successful (LYSA AHL2011, RATHL trial).29
The real prognostic significance of interim PET/CT in HL is yet to be completely validated; thus, patients would probably benefit from the inclusion of additional strategies to complement the therapeutic strategy of PET indication. Decazes et al27 prospectively analysed the correlation between cfDNA and PET/TC parameters in 50 patients with cHL. In particular, two burden parameters as total lesion glycolysis (TLG) and total MTV presented the highest correlation with ctDNA. Among the dispersion parameters, TumBB (which reflects the dispersion volume of the tumours) was highly correlated with ctDNA in cHL. PET/ctDNA correlation is nowadays a growing trend.27
5 LIQUID BIOPSY IN CHL: PREDICTION FOR CLONAL EVOLUTION
Assessment of cfDNA may help to identify clonal evolution in cHL. The panel used by Camus et al was likely too narrow and insufficiently sensitive to detect certain subclonal mutations of low allelic frequency, close to the noise threshold of the NGS sequencer (variant allele frequency [VAF] 0.1%). In addition, it was insufficient for detecting somatic alterations in all patients due to the limited number of included genes.21 However, Spina et al also genotyped longitudinal ctDNA samples from 13 patients collected at diagnosis, at time of relapse, and during salvage therapies. In all cases they pointed out clonal shifts between pretreatment and relapse samples. In particular they inferred that an ancestral clone persisted during the disease course and was characterised by mutations of STAT6, GNA13, ITPKB, and TNFAIP3.20
In a recent report, cfDNA was assessed in patients with R/R HL to characterise the mutational profile and to identify patterns of response or resistance to checkpoint inhibitors.30 Longitudinal samples from 20 R/R cHL patients treated with anti-PD-1 monoclonal antibodies were collected. Blood samples were profiled by the CAPP-Seq strategy. Plasma and peripheral blood mononuclear cells were collected at baseline, every five cycles during therapy administration, and at EOT. Genes recurrently affected by non-synonymous somatic mutations in >30% of R/R cHL patients included STAT6 (50%), SOCS1 (50%), TNFAIP3 (45%), KMT2D (45%), ITPKB (40%), and GNA13 (35%). The longitudinal tracking of ctDNA mutations in R/R cHL patients identified two different patterns of clonal evolution associated with sensitivity (clonal reshaping) or resistance (clonal persistence) to checkpoint blockade. Notably, both patterns correlated with the FDG-PET scan results.30
These data demonstrate that in refractory diseases, chemotherapy only partially reshapes the subclonal composition and cfDNA is a useful tool for monitoring clonal evolution during the therapy of HL.
In conclusion, a combined approach with a useful and harmless tool, the liquid biopsy, is appealing and needed in order to evaluate residual disease that may integrate FDG-PET results for the early identification of chemorefractory patients and to create tailored treatment strategies for cHL patients.
AUTHOR CONTRIBUTION
Conceptualization: F.P., C.G., F.P., G.T. and E.V.; resources: F.P., C.G., G.A., G.R, L.P. and D.L.; writing—original draft preparation: E.V., M.P., F.P. and G.T.; writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
FUNDING INFORMATION
The authors have not received a specific grant for this review from any funding agency in the public, commercial or not-for-profit sectors.
CONFLICT OF INTEREST STATEMENT
Giancarlo Troncone reports personal fees (as speaker bureau or advisor) from Roche, MSD, Pfizer, Boehringer Ingelheim, Eli Lilly, BMS, GSK, Menarini, AstraZeneca, Amgen, and Bayer, unrelated to the current work. The other authors have nothing to disclose.
Open Research
DATA AVAILABILITY STATEMENT
All data relevant to the study are included in the article.