Phylogenomic characterization of the 2022 outbreak of monkeypox virus—The importance of sustained genetic surveillance
Olivia Monteiro and Yan Wa Li contributed equally to the work.
Monkeypox cases are steadily increasing worldwide. Phylogenetic characterization of the monkeypox virus (MPXV) responsible is important for epidemiological studies. Isidro et al. performed shotgun metagenomics sequencing analysis of MPXV isolated from cases in the current outbreak, published in Nature Medicine.1 The results revealed for the first time that these samples cluster with a lower fatality clade of MPXV and that the current outbreak had a common origin. Furthermore, the study found evidence of accelerated evolution in the samples. This study highlights the importance of timely and sustained sequencing efforts to track the potential evolutionary trajectory of MPXV to mitigate its potential impact on global health.
The COVID-19 pandemic and other severe infectious disease outbreaks in recent years have in common among them the ability to cause infections across borders, likely facilitated by air travel and our changing relationship with rural areas.2 The recovery of cross-country travel following the COVID-19 slowdown increases the possibility of bringing predominantly endemic infectious diseases to naïve populations. Reports of monkeypox around the world may be an example and potentially represents the next infectious disease challenge. The first case was recorded on May 6, 2022 in the United Kingdom.3 As of June 27, 2022, over 4300 laboratory-confirmed monkeypox cases have been recorded across six continents.4 We grouped daily new cases into weekly bins (Figure 1A), and it is clear that the number of cases is showing a persistent increase.
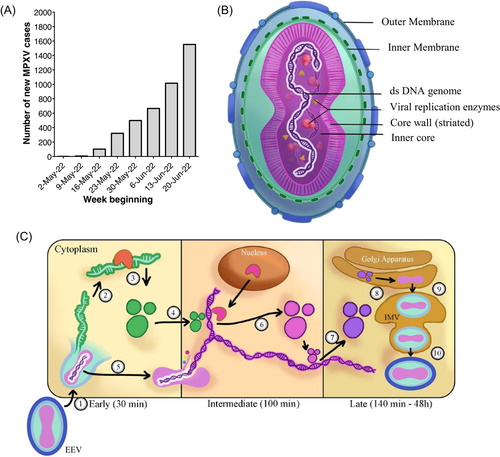
Monkeypox is caused by MPXV, a group of linear double-stranded DNA viruses part of the Orthopoxvirus genus.5 Notable Orthopoxviruses include the Variola virus, the causative agent of smallpox and Molluscum contagiosum. All Orthopoxviruses are morphologically similar, with a brick-like structure (Figure 1B). The genome of Orthopoxviruses contains several hundred nonoverlapping open reading frames (ORFs). Many of these ORFs are highly conserved among members and are required for replication and morphogenesis. Others are divergent and result in heterogeneity in the host range, immune modulation properties, and pathogenesis. All Orthopoxviruses replicate in the cytoplasm of infected cells. Their life cycle is illustrated in Figure 1C.
Monkeypox has a variable incubation period (5–21 days), and is a self-limiting disease with symptoms lasting between 2 and 4 weeks.6 The disease can be divided into two phases. The invasive prodromal phase is characterized by generalized systemic symptoms, such as fever, headache, lymphadenopathy, fatigue, and myalgia. The characteristic monkeypox rash is a feature of the cutaneous phase, which begins 1–3 days after fever onset. These are well-circumscribed, deep-seated, and painful maculopapular skin lesions, which typically occur on the face and then spread to the extremities. Complications of monkeypox are rare, and usually secondary to bronchopulmonary or corneal infections, leading to sepsis, encephalitis, and loss of vision.
MPXV is a zoonotic infection, endemic in more deprived communities in Central and Western Africa. MPXV can be separated into three clades6: Clade 1 (Central African or Congo Basin clade) and Clade 2/3 (collectively West African clade). Clade 1 has a case-fatality ratio (CFR) of 10%, while Clades 2 and 3 have a CFR of <1%. Before the current outbreak, cases of MPXV infections have known travel links to endemic areas or contact with infected animals. However, with the exception of the first confirmed case in the current outbreak (in the United Kingdom), most cases had not been to endemic areas, or contacted the index case. Furthermore, in the current outbreak cases of genital lesions occurring immediately after the prodromal phase have been reported in individuals with close contact with infected individuals, including men who have sex with men, highlighting a sexual route of infection.7 This atypical epidemiology necessitates an analysis of the phylogenetic placement and evolutionary trends of the current MPXV outbreak. The phylogenomic analysis of MPXV reported by Isidro et al. is the first study to offer clues into the genetics of the MPXV strain responsible for the current outbreak.1
This study is based on 15 clinical MPXV samples obtained from lesions and vesicle swabs. The samples were subjected to shotgun metagenomics sequencing and mapped to a reference genome (MPXV-UK_P2, 2018; GenBank accession #MT903344.1). A phylogenetic analysis was performed, using Zaire-96-I-16 (RefSeq accession #NC_003310.1) as a reference sequence for Clade 1 of the global phylogeny, which revealed that these MPXV samples belong to the less virulent Clade 3. Further analysis of other MPXV sequences from NCBI (until June 15, 2022) showed that the current outbreak cluster (lineage B.1.1) forms a divergent branch from lineage B.1, which caused a large outbreak in Nigeria in 2017/2018. Furthermore, the current outbreak cluster differed from the most recent cluster (outbreak in 2018/2019) by 50 single-nucleotide polymorphisms (SNPs). The authors speculated that the current MPXV cluster originated from the virus that caused the 2017/2018 outbreak in Nigeria and continued to evolve. The epidemiological finding of cases in multiple countries within 3 weeks of the index case in the UK suggests that there may be more than one single origin.
Orthopoxviruses generally accumulate 1–2 SNPs per site per year, which means that the current cluster has 6- to 12-fold more SNPs than predicted.8 Isidro et al. suggested that this may be due to accelerated evolution and hypothesized that this may be mediated by human apolipoprotein B mRNA-editing catalytic polypeptide-like 3 (APOBEC3) enzymes, which edit viral genomes to inhibit their replication and infectivity. Microevolution analysis focused on GA > AA and TC > TT replacements, a signature of APOBEC3-mediated genome editing. They found 26 GA > AA and 15 TC > TT replacements in the current MPXV cluster, resulting in 24 non-synonymous, 18 synonymous, and 4 intergenic replacements. Interestingly, 2 (out of 15) of the clinical samples share a frameshift deletion in a gene that codes for an Ankyrin-rich host range protein, resulting in gene loss. The authors speculated that such gene loss during secondary transmission resulted from microevolution of the virus during human-to-human transmission. Additionally, the study also found non-synonymous intra-patient single nucleotide variants (iSNVs) in genes with known immune modulation functions in five of the samples, suggesting an evolutionary adaptation for efficient replication in humans.
The Isidro et al. study has provided the scientific and medical community the first glimpse into the phylogeny of the current MPXV outbreak. Promisingly, it has been identified that the phylogeny of the current outbreak belongs to the lower CFR Clade 3. However, it has also found evidence of accelerated evolution and genomic heterogeneity within single patients. These findings highlight the importance of continued genetic surveillance of the MPXV variants in the current outbreak. Many of the mutations found were non-synonymous, which may impact MPXV infectivity, transmissibility, host range, and also immune modulation. However, there are also a number of synonymous mutations, which may also impact MPXV function. This is particularly important in the context of recent findings that synonymous mutations are rarely neutral.9 Therefore, there is a need to characterize these mutations in functional studies.
Upgrading the current outbreak to one of international concern may depend on the evolutionary trajectory of MPXV, perhaps toward higher virulence and/or CFR. Unmitigated spread increases the rate at which variants occur, and therefore protecting at-risk groups by vaccination will be a priority. A smallpox vaccine with efficacy against MPXV infection is being offered to at-risk groups in the United Kingdom.10 Furthermore, Tecovirimat is a small-molecule antiviral that has been approved for treating monkeypox in Europe since January 2022. These are vital weapons in our arsenal against this emerging infectious disease threat, and may perhaps put the global medical and scientific community on a more solid footing to deal with MPXV. We also need to draw on our experience with Covid-19 and step up genetic surveillance on MPXV, as highlighted by the Isidro et al. study.
AUTHOR CONTRIBUTIONS
Olivia Monteiro, Yan Wa Li, and Daniel T. Baptista-Hon analyzed the MPXV incidence data and produced the figures, and drafted the manuscript. Daniel T. Baptista-Hon conceived the research and provided overall direction. All authors agreed on the content of the final manuscript.
ACKNOWLEDGMENT
The funding source is not applicable.
CONFLICTS OF INTEREST
Author Daniel T. Baptista-Hon is an editorial board member of MedComm – Future Medicine. Author Daniel T. Baptista-Hon was not involved in the journal's review of, or decisions related to, this manuscript. The remaining authors declare no conflict of interest.
ETHICS STATEMENT
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.




