Automated machine learning with interpretation: A systematic review of methodologies and applications in healthcare
Han Yuan, Kunyu Yu and Feng Xie are contributed equally.
Abstract
Machine learning (ML) has achieved substantial success in performing healthcare tasks in which the configuration of every part of the ML pipeline relies heavily on technical knowledge. To help professionals with borderline expertise to better use ML techniques, Automated ML (AutoML) has emerged as a prospective solution. However, most models generated by AutoML are black boxes that are challenging to comprehend and deploy in healthcare settings. We conducted a systematic review to examine AutoML with interpretation systems for healthcare. We searched four databases (MEDLINE, EMBASE, Web of Science, and Scopus) complemented with seven prestigious ML conferences (AAAI, ACL, ICLR, ICML, IJCAI, KDD, and NeurIPS) that reported AutoML with interpretation for healthcare before September 1, 2023. We included 118 articles related to AutoML with interpretation in healthcare. First, we illustrated AutoML techniques used in the included publications, including automated data preparation, automated feature engineering, and automated model development, accompanied by a real-world case study to demonstrate the advantages of AutoML over classic ML. Then, we summarized interpretation methods: feature interaction and importance, data dimensionality reduction, intrinsically interpretable models, and knowledge distillation and rule extraction. Finally, we detailed how AutoML with interpretation has been used for six major data types: image, free text, tabular data, signal, genomic sequences, and multi-modality. To some extent, AutoML with interpretation provides effortless development and improves users' trust in ML in healthcare settings. In future studies, researchers should explore automated data preparation, seamless integration of automation and interpretation, compatibility with multi-modality, and utilization of foundation models.
Abbreviations
-
- AAAI
-
- annual AAAI conference on artificial intelligence
-
- ACL
-
- annual meeting of the association for computational linguistics
-
- AutoML
-
- automated machine learning
-
- ANN
-
- artificial neural networks
-
- AUPRC
-
- area under the precision recall curve
-
- AUROC
-
- area under the receiver operating characteristic curve
-
- BMI
-
- brain machine interfaces
-
- CNN
-
- convolutional neural networks
-
- ChIP-seq
-
- chromatin immunoprecipitation sequences
-
- DNA-seq
-
- DNA sequences
-
- DNase-seq
-
- DNase I hypersensitive site sequences
-
- DSC
-
- dice similarity coefficient
-
- EMG
-
- electromyogram
-
- ECG
-
- electrocardiogram
-
- FE
-
- feature engineering
-
- fMRI
-
- functional magnetic resonance imaging
-
- GMM
-
- Gaussian mixture model
-
- GAN
-
- generative adversarial network
-
- GAT
-
- graph attention network
-
- GBM
-
- gradient boosting machine
-
- GNB
-
- Gaussian Naive Bayes
-
- GRU
-
- gated recurrent unit
-
- HD
-
- hausdorff distance
-
- HLAN
-
- hierarchical label-wise attention network
-
- ICD
-
- international classification of diseases
-
- IB
-
- information bottleneck
-
- ICLR
-
- international conference on learning representations
-
- ICML
-
- international conference on machine learning
-
- IJCAI
-
- international joint conference on artificial intelligence
-
- IOU
-
- intersection over union
-
- KDD
-
- ACM SIGKDD conference on knowledge discovery & data mining
-
- LDA
-
- linear discriminant analysis
-
- LSTM
-
- long short-term memory
-
- LR
-
- logistic regression
-
- LASSO
-
- least absolute shrinkage and selection operator
-
- MDL
-
- minimum description length
-
- ML
-
- machine learning
-
- MLP
-
- multilayer perceptron
-
- MSE
-
- mean squared error
-
- MNase-seq
-
- micrococcal nuclease digestion with deep sequencing
-
- NeurIPS
-
- annual conference on neural information processing systems
-
- OCT
-
- optical coherence tomography
-
- PRISMA
-
- preferred reporting items for systematic reviews and meta-analyses
-
- PSD
-
- predictive sparse decomposition
-
- PSNR
-
- peak signal-to-noise ratio
-
- PNN
-
- probabilistic neural networks
-
- R-CNN
-
- region convolutional neural networks
-
- RF
-
- random forest
-
- RMSE
-
- root mean squared error
-
- RNA-seq
-
- RNA sequences
-
- RNN
-
- recurrent neural networks
-
- SVM
-
- support vector machine
-
- SHAP
-
- shapley additive explanations
-
- TPOT
-
- tree-based pipeline optimization tool
-
- VAE
-
- variational autoencoder
1 INTRODUCTION
The rapid growth of biomedical big data has led to greater opportunities for the deployment of modern data-driven technologies such as machine learning (ML) [1]. ML techniques have achieved substantial success in processing various types of data and performing diverse tasks in the context of healthcare [2-4]. However, the effective exploitation of healthcare data by ML models necessitates the rigorous configuration of every part of the ML pipeline, which relies heavily on specialized technical knowledge and extensive effort.
To help professionals with borderline expertise in data science to better use ML techniques, Automated ML (AutoML) has emerged as a prospective solution. The objective of AutoML, as defined by Yao et al. [5], is to allow computer programs to replace human tuning in the process of determining all or a part of model configurations while maintaining good performance and high computational efficiency. Configurations in this context refer to all factors that are specified prior to model training and affect the final performance, including input data, feature sets, hyperparameters, and model architectures. Therefore, a complete AutoML pipeline encompasses the automation of data preparation, feature engineering, and model development [6].
AutoML techniques in the ML pipeline cater to various levels of coding proficiency. For example, sophisticated AutoML methods such as NASLib [7], which require advanced programming knowledge, aim to provide greater flexibility for experienced ML engineers. AutoML software packages such as auto-sklearn [8] focus primarily on model development, that is, algorithm selection and hyperparameter optimization, targeting users with mediate coding skills. Additionally, commercial AutoML platforms such as Google Cloud's AutoML system and H2O Driverless artificial intelligence (AI) offer no-coding solutions, featuring user-friendly interfaces and rapid convergence capabilities. Table 1 provides an overview of the toolkits developed by leading companies.
| Company | Toolkit | Modality | Website |
|---|---|---|---|
| Amazon | AutoGluon | Multi-modality | https://auto.gluon.ai/stable/index.html |
| SageMaker | Multi-modality | https://aws.amazon.com/sagemaker/canvas/ | |
| Apple | Create ML | Multi-modality | https://developer.apple.com/machine-learning/create-ml/ |
| Vertex AI | Multi-modality | https://cloud.google.com/vertex-ai?hl=en | |
| IBM | AutoAI | Tabular data | https://www.ibm.com/products/watson-studio/autoai |
| watsonx.ai | Multi-modality | https://www.ibm.com/products/watsonx-ai | |
| Meta | Looper | Multi-modality | https://research.facebook.com/publications/looper-an-end-to-end-ml-platform-for-product-decisions/ |
| Microsoft | Azure machine learning | Multi-modality | https://azure.microsoft.com/en-us/products/machine-learning/automatedml/#overview |
| NVIDIA | TAO | Multi-modality | https://developer.nvidia.com/tao-toolkit |
- Note: The companies are listed alphabetically for ease of reference.
In healthcare, the extensive application of ML significantly amplifies the advantages of implementing AutoML approaches. It enables healthcare professionals with borderline ML knowledge to build high-quality models using a fully automated pipeline [9] and further addresses privacy concerns without sharing data with external ML engineers. AutoML systems effectively fill the gap between the lack of ML expertise among healthcare practitioners and the need for data analytics based on ML models [10]. AutoPrognosis [11] describes an end-to-end diagnosis and prognosis modeling framework that helps healthcare professionals leverage clinical data for risk prediction across diverse clinical settings. Additionally, AutoML improves the efficiency of ML engineers by automating tedious and time-consuming tasks such as data preprocessing [12]. For example, nnU-Net [13] introduces a self-configured biomedical image segmentation method that automates the conversion of raw image data into representative structured features.
Although AutoML systems help both healthcare professionals and ML engineers to process medical data effortlessly, the interpretability of these systems should be improved to boost confidence in the reliability of the generated ML models [14]. Given the potentially serious consequences of medical AI failures, greater demands are being placed on the interpretation of ML models in clinical decision-making to fulfill both medical validation and regulatory requirements. Thus, in contrast to conventional AutoML systems primarily centered on ML development, AutoML with interpretation aligns more closely with the real-world requirements in healthcare settings [15].
Because AutoML systems with interpretation are fundamental to facilitating the clinical adoption of AI technologies, we conducted this review to gain insight into how they empower the health community by lowering the entry barrier and enhancing the credibility of ML algorithms. In recent years, several researchers [6, 9, 10, 16-19] have reviewed the development and application of either AutoML or ML interpretations. However, none have provided a systematic and in-depth summary of AutoML with interpretation, particularly its applications in healthcare. In our review, we aim to integrate existing research practices by categorizing data modalities, AutoML techniques, and interpretation methods to acquire a comprehensive understanding of AutoML with interpretation in healthcare and inspire future research topics. The purpose of the categorization is to provide practitioners with an insight into how various AutoML with interpretation systems have been implemented in different medical tasks.
The promising application of AutoML with interpretation in healthcare necessitates a systematic review of cutting-edge research to bridge the gap between technical innovation and practical application. We envisage that this review will empower healthcare practitioners by providing well-organized and referable information about AutoML with interpretation systems, and further facilitate the real-world deployment of ML systems in diverse healthcare settings.
2 METHODS
2.1 Search strategy and data sources
We conducted a systematic review that encompassed both methodology and application studies on AutoML with interpretation for healthcare. We performed a literature search on four databases: MEDLINE, EMBASE, Web of Science, and Scopus. Given that some of the latest ML research is often presented at conferences and may not be included in these four databases, we also searched for research papers in the proceedings of seven relevant and prestigious ML conferences: AAAI, ACL, ICLR, ICML, IJCAI, KDD, and NeurIPS. The searched terms in the medical domain were (“medical” OR “clinical” OR “health” OR “healthcare” OR “medicine”). We also added the terms (“ML” OR “deep learning” OR “AI”) to limit the search to ML-based studies, and (“automated” OR “automatic”) AND (“interpretable” OR “explainable” OR “interpretability”) to include studies on AutoML with interpretation. We restricted our search to papers published before September 1, 2023.
2.2 Inclusion and exclusion criteria
We followed the Preferred Reporting Items for Systematic reviews and Meta-Analyses guidelines [20] to conduct the systematic review. We included all papers published in English that used AutoML with interpretation to perform healthcare tasks. We excluded review articles, workshop papers, duplicate records, and studies not relevant to AutoML with interpretation or healthcare. Each article was independently screened by at least two reviewers, and, if ambiguous, discussed with the corresponding author to reach a consensus.
2.3 Data analysis
Table 2 presents our evaluation and summary of the papers from three aspects: AutoML techniques, interpretation methods, and target data types. For AutoML techniques, we identified three main research directions: automated data preparation, automated feature engineering, and automated model development [9]. For interpretation methods, we summarized from four angles: knowledge distillation and rule extraction, intrinsically interpretable models, data dimensionality reduction, and feature interaction and importance [18, 19]. For target data types, we classified the included articles into six categories: image, free text, tabular data, signal data, genomic sequence, and multi-modality. Additionally, Table 2 lists specific applications and performance advantages of AutoML for users focused on specific tasks.
| Data type | Year | Paper | Automated data preparation | Automated feature engineering | Automated model development | Interpretability methods | Model name | Main ML architectures | Healthcare applications | Performance comparison |
|---|---|---|---|---|---|---|---|---|---|---|
| Image | 2023 | Alkhalaf et al. [21] | √ | √ | Feature interaction and importance | AAOXAI-CD | CNN, RNN, GRU, LSTM | Cancer classification | Accuracy: 99.00/97.00 & 99.42/98.43 | |
| Image | 2023 | Berghe et al. [22] | √ | Feature interaction and importance | U-net, CNN | Structural lesions detection | Accuracy: 0.89/- & 0.92/-, AUROC: 0.92/- & 0.91/- | |||
| Image | 2023 | Cabon et al. [23] | √ | Data dimensionality reduction | LR, RF, SVM | Functional age estimation | MAE: 1.4/- & 1.6/- & 1.3/- | |||
| Image | 2023 | Choi et al. [24] | √ | Knowledge distillation and rule extraction | SimpleMind | CNN, U-net | Endotracheal tube assessment, kidney segmentation, prostate segmentation | Accuracy: 89/-, DSC: 0.881/0.878 & 0.842/0.818, HD: 46.4/30.7 | ||
| Image | 2023 | Custode et al. [25] | √ | Intrinsically interpretable models | U-net, CNN, decision tree | Lung status evaluation | ||||
| Image | 2023 | Dai et al. [26] | √ | Feature interaction and importance | MS-net | CNN | Lung nodules assessment | Accuracy: 92.4/90.0 & 88.5/87.3 | ||
| Image | 2023 | Gerbasi et al. [27] | √ | √ | Feature interaction and importance | DeepMiCa | CNN, U-net | Microcalcifications detection | Accuracy: 0.83/- & 0.83/-, AUROC: 0.95/- & 0.89/-, AUPRC: 0.78/-, IOU: 0.74/- | |
| Image | 2023 | Ghassemi et al. [28] | √ | Feature interaction and importance | GAN | COVID-19 classification | Accuracy: 89.24/88.05 & 98.25/94.69 & 96.20/94.69 & 98.89/96.30 & 99.2/99.6, AUROC: 97.22/96.71 & 99.79/99.03 & 99.43/99.43 & 99.95/99.60 & 99.95/99.99 | |||
| Image | 2023 | Jun et al. [29] | √ | Feature interaction and importance | CNN, U-net | Noninvasive meningioma triaging | AUROC: 0.770/0.757, DSC: 0.910/0.907 | |||
| Image | 2023 | Leong et al. [30] | √ | Feature interaction and importance | Decision tree | Lung water content evaluation | AUROC: 0.719/- & 0.756/- | |||
| Image | 2023 | Orton et al. [31] | √ | Data dimensionality reduction | LASSO | Molecular, histopathology and clinical target prediction | ||||
| Image | 2023 | Pham et al. [32] | √ | Feature interaction and importance | U-net, CNN | Human epidermal growth factor receptor-2 classification | F1-score: 0.80/0.81 | |||
| Image | 2023 | Saglam et al. [33] | √ | Feature interaction and importance | XGBoost, SVM | Early onset schizophrenia classification | Accuracy: 0.80/0.78, AUROC: 0.85/0.83 | |||
| Image | 2023 | Taşcı et al. [34] | √ | Feature interaction and importance | DGXAINet | CNN, SVM | Brain tumor classification | Accuracy: 98.42/95.75 & 99.96/98.91 | ||
| Image | 2023 | Wang et al. [35] | √ | Feature interaction and importance | CNN, U-net | Parkinson's disease classification | AUROC: 0.901/0.856 | |||
| Image | 2023 | Xiang et al. [36] | √ | Feature interaction and importance | CNN, GCN | Prostate cancer classification | Accuracy: 0.677/0.584, AUROC: 0.985/- & 0.986/- | |||
| Image | 2023 | Yoon et al. [37] | √ | Feature interaction and importance | CNN | Anterior disc displacement classification | AUROC: 0.985/0.910 & 0.960/0.861 | |||
| Image | 2022 | Yu et al. [38] | √ | Feature interaction and importance | CNN, RF | Idiopathic pulmonary fibrosis prediction | AUROC: 0.987/- | |||
| Image | 2022 | Basso et al. [39] | √ | Data dimensionality reduction | LDA, LR, RF | Glomerular disorder classification | Accuracy: 77/- & 87/- | |||
| Image | 2022 | Chen et al. [40] | √ | √ | Intrinsically interpretable models | R-CNN, U-net, LR | Blunt splenic injury triaging | Accuracy: 92/-, AUROC: 0.83/0.88 | ||
| Image | 2022 | Falco et al. [41] | √ | Intrinsically interpretable models | Fuzzy rules | COVID-19 classification | Accuracy: 80.67/80.28 | |||
| Image | 2022 | Kakileti et al. [42] | √ | Knowledge distillation and rule extraction | V-net, RF | Early vascularity evaluation | AUROC: 0.85/0.79 | |||
| Image | 2022 | Maqsood et al. [43] | √ | Feature interaction and importance | CNN, SVM | Brain cancer prediction | Accuracy: 97.47/93.85 & 98.92/98.59 | |||
| Image | 2022 | McCay et al. [44] | √ | √ | Knowledge distillation and rule extraction | LR, SVM, decision tree, LDA | Cerebral palsy prediction | Accuracy: 100/100 & 38/38 & 97.37/86.84 | ||
| Image | 2022 | Mou et al. [45] | √ | Feature interaction and importance | DeepGrading | CNN | Corneal confocal microscopy estimation | Accuracy: 84.10/82.40 | ||
| Image | 2022 | Nafisah et al. [46] | √ | Feature interaction and importance | CNN, U-net | Tuberculosis detection | Accuracy: 0.987/0.980, AUROC: 0.999/0.990 | |||
| Image | 2022 | Nijiati et al. [47] | √ | Feature interaction and importance | CNN, U-net | Active pulmonary tuberculosis classification | Accuracy: 0.910/0.895 | |||
| Image | 2022 | Sharma et al. [48] | √ | Feature interaction and importance | CNN, U-net | COVID-19 classification | Accuracy: 97.45/98.70, AUROC: 0.998/0.980 | |||
| Image | 2022 | Park et al. [49] | √ | Feature interaction and importance | U-net, LightGBM | Pilocytic astrocytomas classification | AUROC: 0.930/0.785 | |||
| Image | 2022 | Sharma et al. [50] | √ | Feature interaction and importance | COVID-MANet | CNN, U-net | COVID-19 classification | Accuracy: 97.37/97.16, IOU: 93.64/91.40, DSC: 96.70/95.49 | ||
| Image | 2022 | Suri et al. [51] | √ | Feature interaction and importance | COVLIAS 2.0-cXAI | CNN, U-net | COVID-19 localization | Accuracy: 98.5/98.2, AUROC: 0.990/0.988 | ||
| Image | 2022 | Ullah et al. [52] | √ | Feature interaction and importance | GNB, SVM, decision tree, LR, KNN, RF | COVID-19 classification | Accuracy: 98.5/99.4 | |||
| Image | 2021 | Fu et al. [53] | √ | √ | Feature interaction and importance | CNN, GRU | Brain disease classification | Accuracy: 0.8961/0.9458 | ||
| Image | 2021 | Horry et al. [54] | √ | Intrinsically interpretable models | CNN, decision tree | Lung cancer classification | Accuracy: 0.85/- | |||
| Image | 2021 | Myeongkyun et al. [55] | √ | Knowledge distillation and rule extraction | R-CNN, K-means, SVM | Bacterial pneumonia classification, COVID-19 classification | Accuracy: 91.2/- & 95.0/- | |||
| Image | 2021 | Pietsch et al. [56] | √ | Feature interaction and importance | APPLAUSE | U-net, Gaussian process regression | Placenta health prediction | AUROC 0.95/- | ||
| Image | 2021 | Shorfuzzaman et al. [57] | √ | Feature interaction and importance | CNN | Diabetic retinopathy triaging | Accuracy: 0.962/0.986, AUROC: 0.978/0.997 | |||
| Image | 2021 | Zhao et al. [58] | √ | Feature interaction and importance | LR | COVID-19 classification | Accuracy: 0.9460/0.9249, AUROC: 0.9470/0.9797, DSC: 0.9796/0.9732, HD: 20.2249/41.0517 | |||
| Image | 2021 | He et al. [59] | √ | Feature interaction and importance | CovidNet3D | CNN | COVID-19 detection | Accuracy: 88.69/88.55 & 82.29/81.82 & 96.88/94.27 | ||
| Image | 2021 | Boumaraf et al. [60] | √ | Data dimensionality reduction | CNN | Breast cancer classification | Accuracy: 98.13/87.69 & 98.13/87.69 & 98.26/98.18 | |||
| Image | 2021 | Cheung et al. [61] | √ | Data dimensionality reduction | CNN | Retinal-vessel caliber measurement | ||||
| Image | 2021 | Mosquera et al. [62] | √ | Data dimensionality reduction | CNN | Chest radiography diagnosis | AUROC: 0.7491/- & 0.8745/- | |||
| Image | 2021 | Tamarappoo et al. [63] | √ | Feature interaction and importance | XGBoost | Cardiac events prediction | AUROC: 0.81/0.75 | |||
| Image | 2021 | Yan et al. [64] | √ | Data dimensionality reduction | WBC-Profiler | PSD, RF | Leukocyte classification | Accuracy: 90.22/88.88 | ||
| Image | 2020 | Rucco et al. [65] | √ | Data dimensionality reduction | TPOT, Auto-SkLearn | CNN | Glioblastoma diagnosis | Accuracy: 0.89/0.89 & 0.92/0.89, AUROC: 0.96/0.84 & 0.91/0.84 | ||
| Image | 2020 | Putten et al. [66] | √ | Data dimensionality reduction | CNN | Early neoplasia classification | AUROC: 0.93/0.89 | |||
| Image | 2020 | Wang et al. [67] | √ | Feature interaction and importance | CNN, RNN | Congenital heart disease interpretation | Accuracy: 0.946/0.895 & 0.917/0.878, AUROC: 0.918/0.845 | |||
| Image | 2020 | Yin et al. [68] | √ | Data dimensionality reduction | PNN, SVM, LR, Adaboot, RF, MLP | Bladder cancer diagnosis | Accuracy: 96.7/84.0, AUROC: 0.990/0.926 | |||
| Image | 2020 | Lecouat et al. [69] | √ | Intrinsically interpretable models | Adaptive smoothing, game encoding | fMRI sensing | PSNR: 36.47/37.95 & 44.17/44.09 | |||
| Image | 2019 | Wu et al. [70] | √ | Intrinsically interpretable models | Decision tree | Breast cancer prediction | Accuracy: 72.43/- | |||
| Image | 2019 | Yamamoto et al. [71] | √ | Data dimensionality reduction | Autoencoder, LASSO, ridge regression, SVM | Prostate cancer recurrence prediction | AUROC: 0.884/0.721 | |||
| Image | 2018 | Pereira et al. [72] | √ | Feature interaction and importance | Boltzmann machine, RF | Brain tumor segmentation, penumbra estimation | DSC: 0.84/0.87 & 0.75/0.82 | |||
| Image | 2017 | Song et al. [73] | √ | Data dimensionality reduction | Remurs | LASSO, elastic net | fMRI analysis | Accuracy: 78.15/75.46 | ||
| Free text | 2021 | Diao et al. [74] | √ | √ | Feature interaction and importance | LightGBM | ICD coding | Accuracy: 95.2/91.3 | ||
| Free text | 2021 | Kulshrestha et al. [75] | √ | Feature interaction and importance | Elastic net, XGBoost, CNN | Chest injury prediction | AUROC: 0.93/- | |||
| Free text | 2021 | Blanco et al. [76] | √ | Feature interaction and importance | GRU | Death cause extraction | AUROC: 53.3/52.1 & 49.4/58.8 & 58.2/62.0 | |||
| Free text | 2021 | Dong et al. [77] | √ | Feature interaction and importance | HLAN | GRU | Medical coding | AUROC: 88.4/88.3 & 94.5/96.9 & 88.5/90.2 | ||
| Free text | 2020 | Yang et al. [78] | √ | Feature interaction and importance | AMFF | LSTM | Medical entity tagging | F1-score: 94.48/90.23 & 92.11/88.46 & 68.34/64.61 & 80.51/80.03 | ||
| Free text | 2020 | Li et al. [79] | √ | Feature interaction and importance | MultiResCNN | CNN | ICD coding | F1-score: 0.073/0.068 & 0.608/0.584 | ||
| Free text | 2019 | Atutxa et al. [80] | √ | Feature interaction and importance | RNN, transformer | ICD coding | F1-score: 0.838/0.786 & 0.963/0.935 & 0.952/0.895 | |||
| Free text | 2018 | Duarte et al. [81] | √ | Feature interaction and importance | GRU | ICD coding | Accuracy: 89.320/79.802 & 81.349/70.754 & 76.112/67.404 | |||
| Tabular data | 2023 | Li et al. [82] | √ | Feature interaction and importance | FETCH | MLP | Hepatitis classification | F1-score: 0.9290/0.8839 | ||
| Tabular data | 2023 | Junaid et al. [83] | √ | Feature interaction and importance | SVM, RF, LightGBM | Parkinson's disease prediction | ||||
| Tabular data | 2023 | Islam et al. [84] | √ | Data dimensionality reduction | LR, MLP, RF, XGBoost | Hypertension prediction | AUROC: 0.894/0.829 | |||
| Tabular data | 2023 | Wang et al. [85] | √ | Knowledge distillation and rule extraction | LR | Heart failure prediction | Accuracy: 0.999/0.995, AUROC: 0.981/0.979 | |||
| Tabular data | 2023 | Zhang et al. [86] | √ | Feature interaction and importance | LR, RF, GBM, MLP | Severe acute pancreatitis prediction | Accuracy: 0.910/0.920, AUROC: 0.907/0.849 | |||
| Tabular data | 2022 | Agüero et al. [87] | √ | √ | Knowledge distillation and rule extraction | MLP, GRU, LSTM | Antimicrobial multidrug resistance prediction | Accuracy: 65.40/-, AUROC: 66.73/- | ||
| Tabular data | 2022 | Chou et al. [88] | √ | √ | Feature interaction and importance | XGBoost, RF, LR | Spinal cord injury prediction | AUROC: 0.68/- | ||
| Tabular data | 2022 | Cui et al. [89] | √ | Feature interaction and importance | LR, RF, XGBoost, MLP, GBM | Early death prediction | Accuracy: 0.772/-, AUROC: 0.820/- | |||
| Tabular data | 2022 | Danilatou et al. [90] | √ | √ | √ | Feature interaction and importance | LR, RF, SVM, decision tree | Mortality prediction | AUROC: 0.93/0.85 & 0.87/0.79 | |
| Tabular data | 2022 | Thongprayoon et al. [91] | √ | Feature interaction and importance | RF, decision tree, XGBoost, MLP | Acute kidney injury prediction | Accuracy: 0.72/0.74, AUROC: 0.79/0.78 | |||
| Tabular data | 2022 | Yin et al. [92] | √ | Feature interaction and importance | RF, GBM, MLP, LR, XGBoost | Severe acute pancreatitis prediction | Accuracy: 0.953/0.943, AUROC: 0.945/0.898 | |||
| Tabular data | 2022 | Yu et al. [93] | √ | Feature interaction and importance | XGBoost, LR, GBM, RF, MLP | Mortality prediction | Accuracy: 0.879/0.857, AUROC: 0.888/0.782 | |||
| Tabular data | 2022 | Zhang et al. [94] | √ | Data dimensionality reduction | XGBoost, CNN | Ischemic stroke classification | Accuracy: 0.6020/0.5671, AUROC: 0.6757/0.6532 | |||
| Tabular data | 2021 | Alaa et al. [95] | √ | √ | √ | Knowledge distillation and rule extraction | AutoPrognosis | RF, AdaBoost, MLP | Breast cancer prediction | AUROC: 0.771/0.773 & 0.823/0.792 & 0.777/0.763 & 0.815/0.784 & 0.790/0.778 & 0.803/0.775 |
| Tabular data | 2021 | Chiang et al. [96] | √ | √ | Data dimensionality reduction | RF | Personalized lifestyle recommendations | MAE: 5.34/5.94 & 3.80/4.05, RMSE: 8.24/9.98 & 6.05/6.68 | ||
| Tabular data | 2021 | Laria et al. [97] | √ | Data dimensionality reduction | Deep LASSO | Attention-deficit hyperactivity disorder prediction | RMSE: 0.545/0.561 & 0.494/0.588 | |||
| Tabular data | 2021 | Ikemura et al. [98] | √ | √ | Feature interaction and importance | GBM, XGBoost | Mortality prediction | AUPRC: 0.807/0.736 | ||
| Tabular data | 2021 | Luo et al. [99] | √ | Knowledge distillation and rule extraction | XGBoost | Asthma hospital visit prediction | ||||
| Tabular data | 2020 | Tong et al. [100] | √ | Knowledge distillation and rule extraction | XGBoost | Asthma hospital visit prediction | ||||
| Tabular data | 2020 | Xie et al. [101] | √ | Intrinsically interpretable models | AutoScore | RF | Mortality prediction | AUROC: 0.780/0.778 | ||
| Tabular data | 2020 | Yang at al. [102] | √ | √ | √ | Data dimensionality reduction | mAML | GBM, XGBoost, AdaBoost | Disease classification | |
| Tabular data | 2019 | Senderovich et al. [103] | √ | Feature interaction and importance | Congestion graphs, generalized Jackson networks | Emergency department and outpatient cancer clinic time prediction | RMSE: 36/- & 104/- | |||
| Tabular data | 2018 | Banerjee et al. [104] | √ | Knowledge distillation and rule extraction | RF, LASSO, SVM | Breast cancer prediction | MSE: 0.04/- | |||
| Tabular data | 2018 | Billiet et al. [105] | √ | Intrinsically interpretable models | Interval coded scoring | Elastic net, linear programming | Acute inflammations diagnosis, breast cancer diagnosis, etc. | Accuracy: 0.88/-, AUROC: 0.92383/- | ||
| Tabular data | 2018 | Corey et al. [106] | √ | Data dimensionality reduction | Pythia | RF, LASSO, GBM | Mortality prediction | AUROC: 0.836/- & 0.883/- & 0.916/- & 0.828/- & 0.820/- & 0.781/- & 0.908/- & 0.845/- & 0.890/- & 0.875/- & 0.910/- & 0.850/- & 0.924/- & 0.890/- | ||
| Tabular data | 2018 | Khurana et al. [107] | √ | Intrinsically interpretable models | Transformation graph | Diabetes prediction, oncology prediction | F1-score 0.820/0.615 & 0.895/0.832 | |||
| Tabular data | 2017 | Laet et al. [108] | √ | Data dimensionality reduction | Naïve bayes, LR | Cerebral palsy classification | Accuracy: 91/90 | |||
| Tabular data | 2016 | Drakakis et al. [109] | √ | Intrinsically interpretable models | Decision tree | Histamine H1 receptor binding prediction | Accuracy: 73.21/68.91 & 86.35/84.50 & 70.43/65.12 & 83.60/88.12 | |||
| Tabular data | 2007 | Keles et al. [110] | √ | √ | Intrinsically interpretable models | NEFCLASS | Neuro-fuzzy system | Prostate cancer classification | ||
| Signal | 2023 | Donckt et al. [111] | √ | Intrinsically interpretable models | LR, GBM | Sleep triaging | Accuracy: 0.866/0.864 & 0.831/0.849 & 0.836/0.815 & 0.867/0.875 | |||
| Signal | 2023 | Heitmann et al. [112] | √ | Feature interaction and importance | DeepBreath | CNN, LR | Respiratory pathology detection | AUROC: 0.887/0.703 & 0.739/0.315 & 0.743/0.614 & 0.870/0.896 | ||
| Signal | 2023 | Raeisi et al. [113] | √ | Feature interaction and importance | CNN, GAT | Neonatal seizure detection | AUROC: 0.966/0.957 | |||
| Signal | 2022 | Han et al. [114] | √ | Knowledge distillation and rule extraction | CNN, knowledge graph | Myocardial infarction prediction | Accuracy: 98.88/97.27 & 93.65/93.77 & 94.13/91.54 | |||
| Signal | 2022 | Huang et al. [115] | √ | Feature interaction and importance | Autoencoder | Epilepsy detection | Accuracy: 97.3/72 | |||
| Signal | 2022 | Jahmunah et al. [116] | √ | Feature interaction and importance | CNN | Myocardial infarction detection | Accuracy: 98.9/- & 98.5/- | |||
| Signal | 2022 | Yang et al. [117] | √ | Knowledge distillation and rule extraction | KNN, SVM, RF, MLP | Cardiac abnormalities classification | Accuracy: 99.0/98.7 | |||
| Signal | 2021 | Lee et al. [118] | √ | Feature interaction and importance | CNN | Arrhythmia classification | F1-score: 81.75/82.2 | |||
| Signal | 2021 | Fuchs et al. [119] | √ | √ | Intrinsically interpretable models | Fuzzy rules | Tremor severity assessments | MAE: 1.85/6.41 & 2.30/8.65 | ||
| Signal | 2021 | Kim et al. [120] | √ | Data dimensionality reduction | CNN | BMI channel selection | Accuracy: 76.8/79.6 & 58.3/58.5 & 70.8/71.4 | |||
| Signal | 2019 | Saboo et al. [121] | √ | Data dimensionality reduction | GMM | Active electrodes selection | AUROC: 0.974/0.752 | |||
| Signal | 2019 | Tison et al. [122] | √ | √ | Feature interaction and importance | CNN | Cardiac disease detection | AUROC: 0.94/- & 0.91/- & 0.86/- & 0.77/- | ||
| Genomic sequence | 2021 | Clauwaert et al. [123] | √ | Feature interaction and importance | Transformer | Genome annotation | AUROC: 0.740/0.882 & 0.920/0.961 & 0.976/0.958 & 0.981/0.978 & 0.976/0.964, AUPRC: 0.039/0.035 & 0.057/0.132 & 0.141/0.098 & 0.128/0.128 & 0.141/0.137 | |||
| Genomic sequence | 2020 | Le et al. [124] | √ | √ | Data dimensionality reduction | TPOT-FSS | TPOT, XGBoost | TPOT enhancement | ||
| Genomic sequence | 2019 | Trabelsi et al. [125] | √ | Feature interaction and importance | deepRAM | CNN, RNN | DNA/RNA sequence binding specificities prediction | AUROC: 0.930/- & 0.951/- | ||
| Genomic sequence | 2018 | Nagorski et al. [126] | √ | √ | Data dimensionality reduction | SpaCC | Convex optimization | Cancer epiGenomictics subtype discovery | ||
| Genomic sequence | 2018 | Shen et al. [127] | √ | Data dimensionality reduction | OFSSVM | SVM | Cancer prediction | Accuracy: 82.35/79.41 & 88.24/88.24 & 97.06/97.06 | ||
| Genomic sequence | 2007 | Yap et al. [128] | √ | Knowledge distillation and rule extraction | Bayesian network | Ovarian cancer detection | ||||
| Multi-modality | 2023 | Roest et al. [129] | √ | Feature interaction and importance | CNN, U-net, SVM | Prostate cancer detection | AUROC: 0.81/0.69 | |||
| Multi-modality | 2023 | Wouters et al. [130] | √ | Feature interaction and importance | VAE, LR, cox regression | Cardiac resynchronization therapy outcome prediction | C-statistic: 0.72/0.70 & 0.70/0.72 | |||
| Multi-modality | 2022 | Abbas et al. [131] | √ | √ | Feature interaction and importance | XGBoost, CNN, U-net | Visual acuity prediction | AUROC: 0.849/0.847 | ||
| Multi-modality | 2022 | Gerbasi et al. [132] | √ | √ | Data dimensionality reduction | XGBoost | Stroke prediction | Accuracy: 0.79/-, AUROC 0.85/- | ||
| Multi-modality | 2022 | Gutierrez et al. [133] | √ | √ | Data dimensionality reduction | GA-MADRID | SVM, KNN, decision tree | Alzheimer's disease classification, frontotemporal dementia classification | Accuracy: 0.849/- & 0.872/- & 0.885/- & 0.926/- | |
| Multi-modality | 2022 | Zhang et al. [134] | √ | Feature interaction and importance | Transformer, RF, LSTM | In-hospital mortality prediction, physiological decompensation prediction, length of stay prediction | AUROC: 0.845/0.841 & 0.845/0.826, AUPRC: 0.464/0.453 & 0.180/0.125 | |||
| Multi-modality | 2021 | Ferté et al. [135] | √ | √ | Intrinsically interpretable models | PheVis | LR | Medical condition prediction | AUROC: 0.957/0.994 & 0.987/0.910, AUPRC: 0.798/0.975 & 0.299/0.262 | |
| Multi-modality | 2019 | Li et al. [136] | √ | Feature interaction and importance | KERP | Graph transformer | Medical image report generation | AUROC: 0.686/0.612 & 0.726/0.646 & 0.760/0.689 & 0.862/0.800 | ||
| Multi-modality | 2018 | Chen et al. [137] | √ | Data dimensionality reduction | DASSA | IB, MDL | Disease propagation pattern detection | |||
| Multi-modality | 2017 | Guo et al. [138] | √ | √ | Knowledge distillation and rule extraction | Hierarchical probabilistic framework | Dermatology image analysis | Accuracy: 75.3/62.9, AUROC: 0.78/0.67 |
- Abbreviations: AUPRC, area under the precision recall curve; AUROC, area under the receiver operating characteristic curve; C-statistic, concordance statistic; DSC, dice similarity coefficient; HD, hausdorff distance; IOU, intersection over union; MAE, mean absolute error; MSE, mean squared error; PSNR, peak signal-to-noise ratio; RMSE, root mean squared error.
3 RESULTS
Figure 1 illustrates the literature selection process for this systematic review. Our initial search yielded 2730 papers. We removed 1378 duplicates; hence, we used 1352 records for title and abstract screening. We excluded 1184 records because they were either not relevant to healthcare (n = 331) or did not use AutoML methods (n = 722); were conference papers that were not from listed conferences (n = 9); were not research articles (n = 121); or were not in English (n = 1). As a result, we included 168 articles for full-text review. Finally, we included 118 papers for systematic review. Figure 2 shows the rising trend of publications in AutoML with interpretation for healthcare and indicates that image and tabular data constituted the major subsets for all included publications. In this section, we first summarize AutoML techniques. Then, we elaborate on the ML interpretations used in the included articles. Finally, we summarize the representative AutoML with interpretation systems for different data modalities.
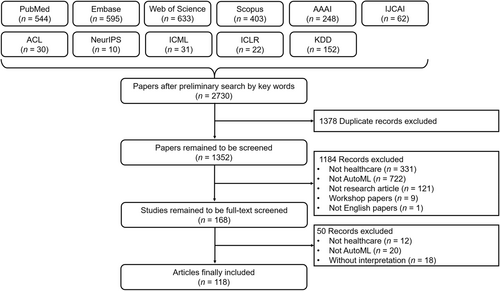
Literature selection flow for automated machine learning with interpretation in healthcare.
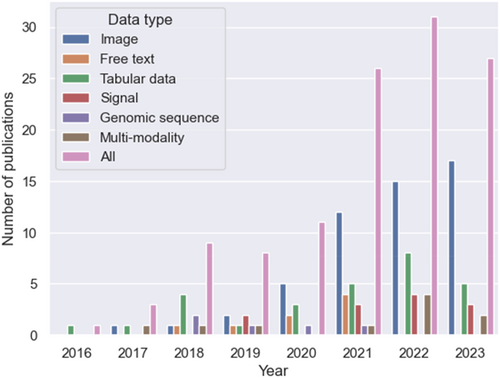
Timeline of publications on automated machine learning with interpretation for healthcare since 2016. Our search concluded on September 1, 2023, which accounts for the lower number of included publications published in 2023 compared with those published in 2022.
3.1 AutoML techniques
For AutoML techniques, we followed the previous classification criteria [9] based on three stages of the ML pipeline: automated data preparation (n = 18), automated feature engineering (n = 95), and automated model development (n = 31). Figure 3 provides a comprehensive overview and Table 3 offers a detailed description of the ML components automated by AutoML within the healthcare sector. Specifically, data preparation refers to the process of collecting and processing raw data into a suitable format for downstream ML stages. AutoML has been leveraged to deal with processes such as automatic data collection [96, 106], noise filtering [27, 28, 44, 119], missing value imputation [87, 95, 110, 126, 133], data imbalance compensation [87, 90, 102, 140], data normalization [44], redundant data removal [53], outlier removal [133], sample clustering [135], data pattern shift detection [137], and continuous variable binning [109]. Feature engineering describes the process of creating new features or modifying existing features to enhance ML performance and AutoML has been used to facilitate automatic feature generation [21, 60, 61, 63, 64, 66, 68, 71, 72, 76, 77, 79-81, 103, 120, 122, 123], selection [70, 72, 80, 98, 99, 101, 102, 105, 108, 121, 124, 127, 135, 141], and transformation [67, 78, 107, 138, 142, 143]. Model development refers to the process of creating, training, and optimizing a model based on either the formatted data or modified features. AutoML has also been used for the selection of main backbone models [65, 86, 88, 89, 91-93, 125, 144], the tuning of model-specific parameters [24, 98, 100, 119, 126, 128, 136], and the optimization of model-specific [21, 24, 40, 59, 74, 86, 89-93, 102, 110, 122, 124, 131, 138, 144] or agnostic hyperparameters [24, 62, 69, 95].
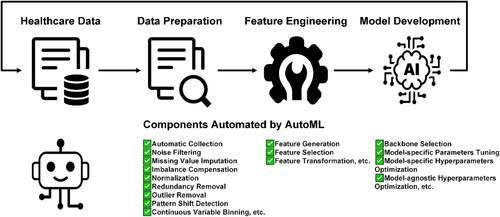
Overview of the ML components automated by automated ML within the healthcare sector. This figure is reproduced from [139] with permission. ML, machine learning.
| Stages | Operations | Description |
|---|---|---|
| Automated data preparation | Automatic data collection | Collecting raw data in an automated manner. |
| Noise filtering | Removing inherent noise from the data. | |
| Missing value imputation | Filling in missing values in the dataset. | |
| Data imbalance compensation | Addressing and compensating for imbalanced classes in the data. | |
| Data normalization | Scaling data to a standard range. | |
| Redundant data removal | Eliminating duplicate or unnecessary data entries. | |
| Outlier removal | Identifying and removing anomalous data points. | |
| Samples clustering | Grouping similar data samples together. | |
| Data pattern shift detection | Detecting changes in data patterns over time. | |
| Continuous variable binning | Converting continuous variables into discrete bins. | |
| Automated feature engineering | Automatic feature generation | Creating features automatically using algorithms. |
| Feature selection | Choosing the most relevant features for modeling. | |
| Feature transformation | Transforming features to a more suitable form for modeling. | |
| Automated model development | Backbone model selection | Choosing the main model architecture. |
| Model tuning | Adjusting model-specific parameters for better performance. | |
| Hyperparameter optimization | Finding the best hyperparameters for better performance. |
Additionally, we conducted a comparative analysis of commonly used metrics between AutoML and the most competitive baseline in the last column of Table 2, which demonstrated that AutoML outperformed conventional ML solutions across various data types. Specifically, slashes (“/”) divide AutoML performance and the most competitive baseline performance. Hyphens (“-”) indicate that specific results were not reported in the original papers. Ampersands (“&”) separate the same evaluation metrics across different tasks or experimental settings and commas (“,”) separate different evaluation metrics. We retained all measurement units and decimal digits from the original papers. We did not report results from studies in which visual performance comparisons were made without quantitative data or from studies involving an excessive number of tasks because of content constraints.
Furthermore, we implemented a toy example to compare AutoML solutions with classic ML models based on 44 918 de-identified patients from BIDMC critical care units [145]. The prediction target was in-hospital mortality (8.81% across all patients) and the candidate variables were age, temperature, platelet, glucose, sodium, lactate, potassium, bicarbonate, heart rate, respiration rate, hematocrit, creatinine, hemoglobin, chloride, anion gap, white blood cells, blood urea nitrogen, systolic blood pressure, diastolic blood pressure, mean arterial pressure, and peripheral capillary oxygen saturation [146]. We randomly divided the entire dataset using the ratio 6:2:2 for model training, validation, and testing. For traditional ML models, we optimized the hyperparameters using grid search based on the area under the receiver operating characteristic curve (AUROC) evaluated on the validation set. For AutoML solutions, we automatically determined the hyperparameters using their inherent algorithms; therefore, their training data included both the training and validation sets. Figure 4 presents the AUROC results on the unseen test set, which demonstrates that the two AutoML solutions of AutoGluon [147] and TPOT [124] statistically significantly outperformed the conventional ML models random forest [148], gradient boosting machine (GBM) [149], and K-nearest neighbor [150]. We made the code open access to enable reproducibility and serve as an exemplary case study [151].
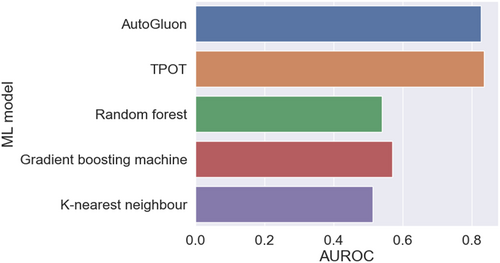
AUROC comparison of automated ML solutions versus conventional ML methods for real-world in-hospital mortality prediction. AUROC, area under the receiver operating characteristic curve; ML, machine learning.
3.2 Interpretation methods
Regarding the ML interpretations, we grouped them into four categories based on the commonly adopted criteria [18, 19]: feature interaction and importance (n = 63), data dimensionality reduction (n = 27), intrinsically interpretable models (n = 14), and knowledge distillation and rule extraction (n = 14).
Feature interaction entails quantifying the effect of one feature on another, considering their mutual influence, whereas feature importance involves discerning the significance of input features in shaping the output targets of ML models [61, 63, 67, 72, 76, 78-81, 98, 103, 108, 122, 125, 136]. In the healthcare domain, the alignment of feature interaction and importance with clinical expertise enhances healthcare professionals' trust in ML outputs [152]. However, when feature interaction and importance diverge from established knowledge, ML models may encounter overfitting issues. Remarkably, such disparities occasionally reveal previously unidentified biomarkers [153].
Data dimension reduction refers to the use of a subset of the most informative raw inputs or modified features in model development and subsequent analyses [61, 62, 64-66, 68, 71, 73, 76, 102, 106, 108, 120, 121, 124, 126, 127, 137]. In the context of high-dimensional samples, data dimension reduction helps the model to focus on salient features, thereby simplifying model complexity and enhancing its interpretability [154]. Additionally, data dimension reduction enables the effective graphical visualization of data distributions within a low-dimensional space [155]. This visualization reveals latent data patterns that can be integrated into subsequent model development, thereby enhancing both model performance and interpretability [18, 19].
Intrinsically interpretable models represent the application of transparent models to solve prediction problems [18] such as logistic regression [101, 111, 135, 140, 141, 156, 157], decision tree [25, 54, 70, 105, 137], fuzzy rules [41, 110, 119], and mathematical solid decision functions [40, 69, 107]. Intrinsically interpretable models feature simple architectures or algorithms, thereby fostering a clear understanding of the relationship between inputs and outputs [18, 19]. These models may not consistently achieve predictive performance comparable with that of their black box counterparts, but within high-stakes tasks that impact lives, model transparency is substantially more important than marginal performance superiority [158].
Knowledge distillation and rule extraction refer to the processes of simplifying intricate ML models into either streamlined models or human-comprehensible rules, respectively [99, 100, 104, 128, 138]. Knowledge distillation is a technique designed to train simple student models by mirroring the behavior of complex teacher models while preserving model performance [159]. Post distillation, student models demonstrate reduced complexity, which renders them more comprehensible to humans and potentially bolsters transferability [160]. Rule extraction yields human-understandable rules because each rule inherently provides a logical explanation for its decision [161]. Based on these interpretation methods discussed above, healthcare practitioners can discern potential errors and ascertain the reliability of ML models [162].
3.3 Data modalities
In this section, we discuss AutoML with interpretation for different types of healthcare data: image (n = 53), free text (n = 8), tabular data (n = 29), signal (n = 12), genomic sequence (n = 6), and multi-modality (n = 10). Figures 5 and 6 present the summary statistics of AutoML and interpretation techniques in the included publications. For AutoML techniques, automated feature engineering dominated in five out of the six modalities; for the genomic sequence, automated model development was more prevalent. Regarding interpretation methods, feature interaction and importance were widely used in all modalities except the genomic sequence, where data dimensionality reduction was the preferred approach. For each data modality, we focused on the principal tasks addressed by AutoML with interpretation systems and elaborated on them using representative studies.
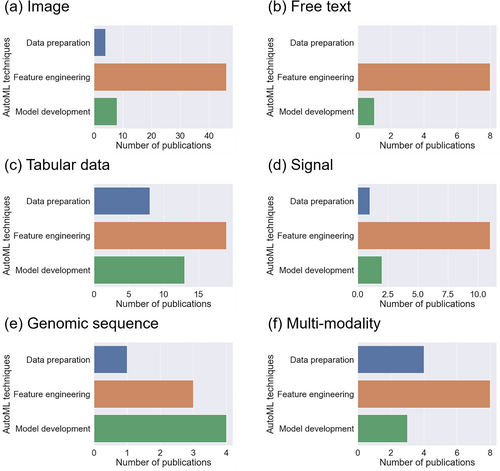
Summary statistics of automated machine learning techniques for the included publications targeting each data modality: (a) image; (b) free text; (c) tabular data; (d) signal; (e) genomic sequence; and (f) multi-modality.
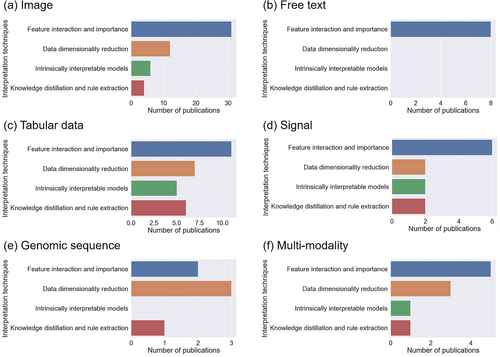
Summary statistics of interpretation techniques for the included publications targeting each data modality: (a) image; (b) free text; (c) tabular data; (d) signal; (e) genomic sequence; and (f) multi-modality.
3.3.1 Image
Medical images are essential diagnostic tools for a spectrum of diseases [66, 163]. AutoML with interpretation enables clinicians with little coding experience [164] to perform a spectrum of healthcare tasks, such as retinal-vessel caliber measurement [61], breast cancer classification [60], and thoracopathy lesion localization [165]. Based on whether they transform raw pixels into useable features, current systems can be classified into two categories: (1) two-step systems that consist of feature extraction and subsequent modeling [64, 66, 68, 70, 71, 166]; and (2) end-to-end systems without the explicit extraction of intermediate features [67, 69].
Two-step systems first extract image features from raw pixels and build up the subsequent analysis based on the extracted features. Various methods have been proposed to automate the extraction of image features, including both commercial software and homemade models. Yin et al. [68] applied the commercial software CellProfiler [167] and ImageJ [168] to extract individual and textual features, and then integrated domain knowledge from pathologists to shortlist useful features. PDE [66] has also demonstrated its effectiveness in automatic feature extraction. By contrast, Yan et al. [64] developed a feature extraction tool and demonstrated the effectiveness of their homemade model through a comparison with human clinicians. With diverse off-the-shelf solutions, multiple tools have been combined to improve the robustness of extracted features [169]. In these systems, the most common interpretation is feature interaction and importance that results from mapping the extracted features back to the original images and highlighting relevant pixels or patches [64, 71]. Additionally, in some systems, inherently interpretable models are applied based on the extracted features to improve model interpretations [70, 166]. For instance, Wu et al. [70] implemented a decision tree to mimic how radiologists interpret the extracted features. Moreover, knowledge distilled from an inherently interpretable model, such as a decision tree, can serve as diagnostic guidelines in the future [166].
Different from two-step methods, end-to-end systems process image inputs without the implicit extraction of intermediate features and output predictions of interest in addition to useful interpretations [170]. In the task of compressed sensing for functional magnetic resonance imaging (fMRI), Lecouat et al. [69] automated the architecture design and parameter training of artificial neural networks (ANN) based on convex optimization and non-cooperative games [171]. To enhance interpretability, they introduced a decision function with sparse parameters and clear mathematical formulas. Wang et al. [67] developed a classic end-to-end system for congenital heart disease classification, including automatic data clustering and model parameter tuning. Similar to two-step systems, their system highlighted important areas on the input image toward ML predictions and used these sub-areas as an interpretation. Although end-to-end systems provide more ceaseless automation and are thus more user-friendly, users should choose the appropriate systems based on whether they need the intermediate features for further modeling and interpretation [68].
3.3.2 Free text
Medical text records various patients' information, such as hospitalization descriptions, diagnoses, and treatments [172]. Accurate mining of such information can summarize patients' former health conditions and guide subsequent interventions [173]. A fundamental task addressed by AutoML with interpretation is the coding of unstructured raw clinical notes into structured medical codes, such as the international classification of diseases (ICD). Similar to the two-step systems adopted in medical image analysis, this process extracts standard intermediate features from text records, and these intermediate features facilitate various subsequent analyses [80]. Conventionally, such a transformation was conducted manually [79, 80], but has been gradually replaced by either commercial software or home-made models to save time and eliminate errors. For example, commercial software called clinical text analysis and knowledge extraction system has been demonstrated to be an effective method for mapping trauma encounter text to structured medical concepts [75]. Additionally, researchers have demonstrated that homemade models are useful for generating informative feature vectors from free text and subsequently projecting these vectors to medical codes [74, 76, 77, 80, 81]. Duarte et al. proposed a framework similar to residual learning, wherein word embeddings are processed using a gated recurrent unit (GRU) to generate representations [81]. These representations are then concatenated with the initial embeddings to prevent information loss and enhance model accuracy. Additionally, Atutxa et al. demonstrated that beyond classic recurrent neural networks (RNN) such as GRU, convolutional neural networks (CNN) and transformers are also effective for mapping diagnostic text to ICD codes [80].
Across all analytical tasks that use medical text, the attention mechanism is the most important backbone. It is valued not only because of its superior performance in the attention-based transformer [174] but also because of its inherent weights that provide feature interaction and the importance of each part in the input text [76, 78, 81]. For instance, in the sentence “He should undergo chemotherapy when he is diagnosed before his cancer cells metastasize,” attention detects that “his cancer cells metastasize” is a crucial component in the automatic determination of the patient's cause of death [76]. In recent studies, researchers introduced the hierarchical attention mechanism, which uses the various types of attention and interprets feature representations on different levels. The hierarchical label-wise attention network [77] applies two-level attention mechanisms at the word-level and sentence-level for selecting important words and sentences in each paragraph, respectively.
3.3.3 Tabular data
Tabular data, the most common data format in healthcare, includes structured demographic data, vital signs, lab tests, diagnoses, treatments, and procedures [1]. Unlike pixels in images and words in free text, raw features, such as gender in tabular data, typically have clinically explainable meanings, therefore feature engineering becomes the focal point of automation and interpretation for AutoML systems. It should be noted that the proposed methods for tabular data in the included studies can also be applied to structured information derived from unstructured healthcare data, as illustrated in the two-step methods above. In this section, we focus on studies in which researchers explored raw inputs in a structured tabular format.
Traditional feature engineering for tabular data is labor-consuming and costly. It requires ML engineers' intuition and domain knowledge [107]. By contrast, automatic and interpretable feature engineering automatically performs transformation and aggregation across candidate features in a transparent manner. For example, Khurana et al. [107] proposed automatic feature selection and transformation based on intrinsically interpretable transformation graphs, and found that the modified features reduced ML errors. Their work demonstrated the utility of intrinsically interpretable models in feature engineering. AutoScore [141, 156] further exploits the full potential of the intrinsically interpretable clinical score as the backbone for predicting parameters such as the in-hospital mortality rate [141, 156], survival time [157], and rare event occurrence [140]. Although complicated ML models have dominated the analysis of high-dimensional data, for tabular data with a limited number of features, transparent features and intrinsically interpretable models are still preferred in practice [175, 176].
In addition to feature engineering, data preparation (pre-processing) [106] and model development [98] have been automated using AutoML with interpretation systems. Ikemura et al. [98] automated the entire ML lifecycle using commercial software [177] and interpreted models through feature interaction and importance generated by Shapley additive explanations (SHAP) [178]. In addition to commercial software such as H2O.ai, researchers have also developed comprehensive home-made systems for mining clinical tabular data. mAML [102] is an example that includes automated imbalance compensation [179], feature selection [180], and hyperparameter optimization [181]. Specifically, imbalance compensation is addressed using RandomOverSampler [179], SMOTE [182], and ADASYN [183]. Feature selection methods include the distal DBA method [184], HFE [180], and mRMR [185]. Hyperparameter optimization is performed using a grid search [181].
3.3.4 Signal data
Signal data refers to electrical or mechanical signals collected from physiological sensors to monitor the functioning of the human body and make informed intervention decisions [186]. ML has been applied to identify the sophisticated relationships between various signal inputs and clinical events. AutoML with interpretation further automates and improves the reliability of this analytical process. A promising research direction involves transforming signal data into two-dimensional representations and subsequently applying image-related methods [118]. However, in this section, we focus on these techniques specifically designed for signal data to avoid confusion. Specifically, Fuchs et al. [119] used an intrinsically interpretable fuzzy model to analyze tremor signals, in which the wrapper approach [187] and pyFUME [188] automate feature selection and model development, respectively. Kim et al. [120] proposed an automated channel selection method based on CNN for analyzing electroencephalograms. They further elucidated neurophysiological feature interaction and importance by correlating the selected channels with specific brain regions. In addition to the end-to-end architecture, Tison et al. [122] devised a two-step framework for predicting distinct heart diseases. Initially, the system autonomously generated features using a CNN-hidden Markov model from electrocardiograms (ECG). Subsequently, these features were input into a GBM for predicting the target diseases. Finally, the system calculated the interaction and importance of segments within ECG as the model interpretation. A similar strategy was implemented by Jahmunah et al. [116] in which ECG beats were first extracted using an off-the-shelf algorithm and then input into the downstream DenseNet [189] for myocardial infarction detection. Han et al. [114] conducted an extensive investigation into the use of AutoML for diagnosing myocardial infarction. On top of clinical standards, diagnostic guidelines, and DenseNet-based signal morphology, they developed an interpretable diagnostic system based on production rules.
3.3.5 Genomic sequence
Genomic sequence data [190, 191] indicate the precise order and arrangement of fundamental genetic elements, such as nucleotides (adenine, thymine, cytosine, and guanine), within DNA sequences (DNA-seq). In addition to DNA-seq, other common genomic sequences include RNA sequences (RNA-seq), Deoxyribonuclease I hypersensitive site sequences (DNase-seq), micrococcal nuclease digestion with deep sequencing (MNase-seq), and chromatin immunoprecipitation sequences (ChIP-seq). These sequences encapsulate the detailed composition of genetic material, thereby offering fundamental information that is essential for comprehending potential associations between genetic patterns and diseases [192]. The principal application of AutoML with interpretation in genomic sequence data mining is to identify genomic sites of interest from the entire genomic sequence. Trabelsi et al. [125] proposed deepRAM for identifying protein binding sites in DNA and RNA-seq based on a hybrid architecture of CNN and RNN. The hyperparameters were automatically tuned through a combination of random search and cross-validation. Sequence motifs, which represent patterns with biological significance, were extracted from the initial CNN layer to improve interpretability [125]. In addition to genomic sites, AutoML has been applied to the data mining of gene expression data. Shen et al. [127] introduced elastic net-based [193] automatic feature selection to a support vector machine (SVM), which demonstrated that feature selection boosted both model performance and interpretability. In addition to classic ML models such as SVM, the transformer has gradually gained popularity in genomic sequence analyses, such as automatic prokaryotic genome annotation [123]. In addition to inherent attention in the transformer for acquiring feature interaction and importance, data dimensionality reduction [124] and rule extraction [128] are used to improve model interpretability.
3.3.6 Multi-modality
Multi-modality refers to the simultaneous use of more than one data type discussed above to gain a comprehensive understanding of a patient's condition [194]. The integration of these complementary modalities enhances the overall diagnostic accuracy of ML models [195]. AutoML with interpretation is highly valued for processing complex data that involve multiple modalities [196]. PheVis [135] uses a dictionary-based named entity recognition tool to extract medical concepts from free text and then fuses these features with diagnosis codes to predict rheumatoid arthritis and tuberculosis. The SAFE algorithm [197] is used for automatic feature selection, and logistic regression is used for the transparent modeling of the relationship between shortlisted features and medical conditions of interest. Similarly, Zhang et al. [134] combined phenotypical features from free text and clinical features from tabular data to predict in-hospital mortality, physiological decompensation, and length of stay in intensive care units. Compared with features from a single modality of either free text or tabular data, multi-modal features have led to statistically significant improvements in performance across most evaluated settings. For analogous frameworks within the field of image modality and signal modality, readers can refer to Abbas et al. [131] and Wouters et al. [130], respectively. They used different tools to extract features from image or signal data and combined them with tabular features, which achieved state-of-the-art performance. In addition to integrating different data modalities for predicting events of clinical interest, the aligned data of different modalities facilitates the translation of high-dimensional data into human-understandable formats, such as human language. KERP [136] was proposed to automatically generate free text reports for medical images, where feature interaction and importance, derived from attention weights, are leveraged to connect generated reports with original image regions, mimicking the inference process of a human radiologist.
In addition to the six detailed data categories above, healthcare data can also be generally classified as spatial or sequential data. Image data primarily encompasses spatial information, whereas temporal tabular data, free text, signal data, and genomic sequence data fall into the sequential data category. Medical videos represent an integration of both spatial and sequential data. The shared characteristics across different modalities pave the way for a unified architecture that is capable of handling various data types. Chen et al. [137] designed DASSA for automatic pattern change detection within any sequential data and demonstrated its potential for analyzing the aforementioned sequential data within a unified framework.
4 DISCUSSION
As a fundamental component for the successful implementation of ML in healthcare, AutoML with interpretation reduces the barriers to the full lifecycle of ML analyses and provides interpretations for healthcare professionals [198]. Through a systematic literature review, we discussed the methodologies and applications of AutoML with interpretation for six data types: image, free text, tabular data, signals, genomic sequence, and multi-modality. We identified three components that have been automated in ML analyses: data preparation, feature engineering, and model development. We summarized four major interpretation methods: feature interaction and importance, data dimensionality reduction, intrinsically interpretable models, and knowledge distillation and rule extraction. Using Table 2, readers can easily identify papers in which AutoML with interpretation and model performance are discussed for their tasks of interest. Despite the promising performance achieved by AutoML with interpretation systems, several challenges persist, including the absence of automatic data preparation, the loose integration of automation and interpretation, and the unmet compatibility with multi-modality. Additionally, the latest advancements in foundation models have the potential to revolutionize AutoML with interpretation.
The first challenge of current AutoML with interpretation systems is the absence of automatic data preparation, as highlighted by the finding that automatic data preparation was integrated into AutoML with interpretation systems in only 18 out of 118 studies [199]. Real-world healthcare records contain issues such as missing values, outliers, inconsistencies, duplicates, and non-standardization [200]. These issues constitute almost 50%–80% of the overall workload in the complete lifecycle of ML analyses, underscoring the necessity for automated data preparation within the infrastructure of future AutoML systems [201]. Additionally, we suggest that ML engineers should frequently communicate with healthcare professionals during the system design phase to align their work with real-world demands [202]. For instance, although complex ANNs have become the primary choice in some application domains, such as reinforcement learning [203], intrinsically interpretable models are favored in healthcare settings, such as emergency departments [204]. Hence, ML engineers should ensure the inclusion of common intrinsically interpretable models in their systems rather than exclusively incorporating various ANN architectures.
The second challenge identified in the included papers is the loose integration of automation and interpretation. In all the included studies, the researchers addressed interpretation issues to some extent. Researchers should leverage the insights gained from interpretation to enhance their model automation rather than merely adding post hoc explanations as the last module in their frameworks. A good demonstration was provided by Ikemura et al. [98]. They applied SHAP and PD plots to analyze the decision processes of their AutoML models, indicated potential medical knowledge from their studies, and further reused these findings to enhance their models. The interaction between model development and model interpretation can be achieved by automated feature selection, which reveals feature importance, offers model interpretation, simplifies model structure, and potentially enhances model performance [205]. In addition to automated feature selection, for future AutoML with interpretation, researchers should explore the research direction of developing the tightly knit integration of AutoML and ML interpretations.
Furthermore, the expanding collection of multi-modalities presents an opportunity for ML engineers to develop an AutoML with interpretation system that emulates a human clinician's inference process based on various types of healthcare data [177]. Specifically, when patients visit a hospital, clinicians and nurses investigate their former medical records, which are in the form of text and tabular data. Then, some tests may be conducted on the feedback image and signal data. Some advanced treatments involve genome sequencing, which introduces genetic data into the consultation and diagnosis. Handling such abundant and complex information requires a great deal of domain knowledge. The scenario becomes even more intricate when healthcare professionals seek to leverage ML, and this is an exact application scenario for AutoML with interpretation systems. Given the recent versatile application of the transformer for the data types image [206], free text [207], signal data [208], and genomic sequence [209], future researchers can explore the development of comprehensive AI doctors that use multi-modal healthcare data as inputs, automate the entire pipeline of data analyses, and generate results along with interpretations based on a unified backbone architecture.
Recent advancements in foundation models for text, image, and multi-modality have the potential to significantly enhance all three stages of ML: data preparation, feature engineering, and model development [210]. These models excel in zero-shot learning, which enables them to perform tasks without additional training on specific datasets. For example, large language models, such as ChatGPT, can perform a range of tasks from ICD code extraction [211] to risk triage prediction [212] based on prompts provided by healthcare professionals. This zero-shot capability elevates ML to an unprecedented level of automation, potentially obviating the need for tedious data preparation and computationally intensive model development in certain tasks [213]. By contrast, in tasks in which foundation models exhibit suboptimal performance, they can serve as effective tools for feature engineering. The representations within their architectures can be extracted to enhance downstream models [214]; in previous studies, researchers validated that downstream models embedded with these representations outperformed powerful baseline models [215].
Our study had certain limitations that warrant refinement in future work. First, we sought to provide an overview of current AutoML with interpretation systems in healthcare settings. Hence, we did not consider the technical details of AutoML and interpretation techniques. For readers interested in these technical intricacies, we recommend referring to the original papers for a more in-depth exploration. In future work, we may conduct a detailed review of areas such as the underlying algorithms, methodologies, and implementation frameworks. Second, for a given data modality, various commercial software and homemade solutions are readily available, as illustrated above. Although Figure 4 exemplifies the effectiveness of AutoML in predicting in-hospital mortality for a real-world application, we refrained from suggesting a one-size-fits-all solution because of the heterogeneous properties of datasets across different scenarios. In future endeavors, we could undertake a thorough benchmarking analysis to delineate guidelines. An exemplary precedent is in the investigation conducted by Gijsbers et al. [216], wherein they meticulously scrutinized 9 AutoML frameworks across 71 classification and 33 regression tasks. Finally, to ensure that all the reviewed papers underwent peer review, we excluded preprints, which may have resulted in the latest developments in the field being overlooked. In future studies, we could explore the integration of bibliometric methodologies to discern high-quality preprints from a broader pool, thereby enhancing the comprehensiveness of paper inclusion [217].
5 CONCLUSION
AutoML with interpretation is essential for the successful uptake of ML by healthcare professionals. This review provides a comprehensive summary of the current state of AutoML with interpretation systems in the context of healthcare. To some extent, the proposed systems facilitate effortless development and improve users' trust in ML in healthcare settings. In future studies, researchers should focus on automated data preparation, the seamless integration of automation and interpretation, compatibility with multi-modalities, and the utilization of foundation models to expedite clinical implementation.
AUTHOR CONTRIBUTIONS
Han Yuan: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); visualization (equal); writing—original draft (lead); writing—review & editing (lead). Kunyu Yu: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); writing—original draft (equal); writing—review & editing (equal). Feng Xie: Conceptualization (equal); data curation (equal); investigation (equal); methodology (equal); writing—original draft (equal); writing—review & editing (equal). Mingxuan Liu: Formal analysis (supporting); investigation (supporting); writing—original draft (supporting); writing—review & editing (supporting), Shenghuan Sun: Formal analysis; investigation; writing—original draft.
ACKNOWLEDGMENTS
We would like to acknowledge Prof. Nan Liu at Duke-NUS Medical School for his invaluable support. Additionally, we thank Dr. Ahmed Allam and Prof. Michael Krauthammer at the University of Zurich for their helpful discussions. The funder played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONFLICT OF INTEREST STATEMENT
All authors declare that they have no conflicts of interest.
ETHICS STATEMENT
This study is exempt from review by the ethics committee because it does not involve human participants, animal subjects, or sensitive data collection.
INFORMED CONSENT
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.




