Associations Between Lipids and Lipid-Lowering Drug Target Genes and Osteomyelitis: A Mendelian Randomization Analysis
Funding: This work was supported by the Tertiary Education Scientific Research Project of Guangzhou Municipal Education Bureau (no. 202235415), the Guangdong Basic and Applied Basic Research Foundation (no. 2019A1515010390), and the Elite Talent Programme Project of The Third Affiliated Hospital of Guangzhou Medical University (no. 110340803).
Zhiyi Zhou and Zhehan Yang contributed equally to this work and share the co-first authorship.
ABSTRACT
Background
Lipid metabolism is a key regulator of inflammation in acute and chronic conditions. However, whether dyslipidemia is related to the process of osteomyelitis remains unclear. This study aimed to use a Mendelian randomization (MR) analysis to examine the associations between triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), and osteomyelitis. Additionally, the associations between the genes corresponding to these traits and osteomyelitis were investigated.
Methods
Genetic variants associated with TG and TC were selected from the Global Lipids Genetics Consortium, while LDL-C datasets were extracted from the UK Biobank. Specifically, the lipid-lowering drug target regions were selected as proxies for drug target perturbation. Osteomyelitis was identified according to the FinnGen consortium. We also conducted supplementary analyses using C-reactive protein genome-wide association study data to examine the effect of drug targets on this inflammatory marker. Furthermore, we conducted mediation analyses focusing on several risk factors for osteomyelitis.
Results
No association was found between LDL-C, TG, or TC concentrations and osteomyelitis. Proprotein convertase subtilisin/kexin type 9 (PCSK9) was significantly associated with a lower risk of osteomyelitis (odds ratio [95% confidence interval] = 0.49 [0.32–0.76], p = 1.60 × 10−3) and a lower concentration of C-reactive protein (0.94 [0.92–0.97], p = 3.16 × 10−4). We found that waist circumference was an intermediate variable between PCSK9 and osteomyelitis.
Conclusions
This study does not support a relationship between dyslipidemia and osteomyelitis. PCSK9 is associated with a lower risk of osteomyelitis. Our findings suggest that waist circumference is a potential mediator between osteomyelitis and PCSK9. Additionally, PCSK9 is associated with reduced CRP concentrations.
Abbreviations
-
- APOB
-
- apolipoprotein B-100
-
- APOC3
-
- apolipoprotein C3
-
- CHD
-
- coronary heart disease
-
- CI
-
- confidence intervals
-
- CRP
-
- C-reactive protein
-
- HMGCR
-
- 3-hydroxy-3-methylglutaryl-CoA reductase
-
- LDL-C
-
- low-density lipoprotein cholesterol
-
- LDLR
-
- low-density lipoprotein receptor
-
- LPL
-
- lipoprotein lipase
-
- PCSK9
-
- proprotein convertase subtilisin/kexin type 9
-
- T2D
-
- type 2 diabetes
-
- TC
-
- total cholesterol
-
- TG
-
- triglyceride
-
- WC
-
- waist circumference
1 Introduction
Osteomyelitis is defined as inflammation caused by a bacterial infection in the bone marrow, bone cortex, and periosteum, and is characterized by necrotic bone and fistulous tracts extending from the skin to the bone due to trauma or bacterial infection [1]. Osteomyelitis commonly occurs in the vertebrae, the feet of patients with diabetes, or sites of penetrating bone injuries caused by trauma or surgery. With an increasing incidence rate, osteomyelitis has become a growing concern in modern medical care, with approximately 22 cases/100,000 people in the United States [2]. As a complex and severe disease, osteomyelitis imposes a substantial medical, economic, and social burden on society and people, which causes enormous healthcare costs and resource consumption.
Lipid metabolism is a critical regulator of inflammation in acute and chronic diseases. Growing evidence suggests that lipids are closely associated with various inflammatory and infectious diseases. In various infections, such as bacterial, viral, tuberculosis, and parasitic infections, alterations occur in plasma lipid concentrations [3]. Therefore, therapeutic strategies targeting lipid metabolism may help reduce inflammation and optimize immune function [4]. However, whether lipid metabolism contributes to osteomyelitis is unknown. The association between disorders of lipid metabolism and osteomyelitis remains unclear.
MR can establish a connection between a risk factor and an outcome while also enabling estimation of the causal effect with reduced confounding and bias. The use of instrumental variables (IVs), usually represented by single nucleotide polymorphisms (SNPs), can infer a causal relationship between an exposure and outcome by simulating a randomized, controlled trial environment [5]. MR can be divided into “biomarker MR” and “drug target MR.” In biomarker MR, the association between exposures and outcomes can be investigated. Drug target MR can be used to investigate the effect of genetic variations encoding therapeutic drug targets on related biological effects that are similar to the effect of therapeutic drugs on target genes through intervention mechanisms, affecting their expression or function [6].
Lipid metabolism disorders are related to the development of inflammation. Therefore, we used Mendelian randomization (MR) to investigate the associations between three lipids, triglyceride (TG), total cholesterol (TC), and low-density lipoprotein cholesterol (LDL-C), and osteomyelitis. Furthermore, we examined the associations between lipid-related drug target genes and osteomyelitis to investigate the relationship between lipid-lowering drug targets and osteomyelitis. We also used relevant secondary indicators and examined related pathways to support the relationship between osteomyelitis and the lipid drug target.
2 Methods
This study followed the Mendelian randomization reporting guidelines for enhancing the reporting of epidemiological observational studies [7] (Supporting Information S1: Table S1). Figure 1 shows a flowchart of the research design. Supporting Information S1: Tables S2 and S3 show the details of selecting all instrumental variables (IVs).
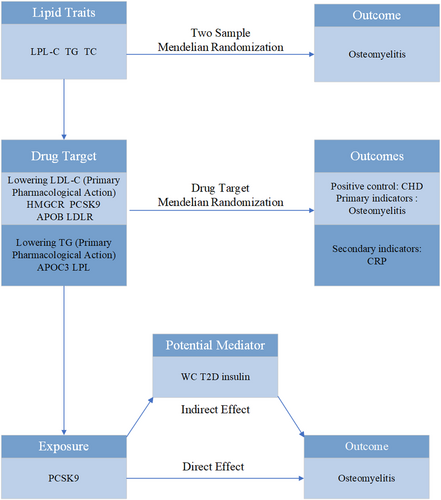
Flowchart of the study design. APOB, apolipoprotein B-100; APOC3, apolipoprotein C3; CHD, coronary heart disease; CRP, C-reactive protein; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; LDL-C, low-density lipoprotein cholesterol; LDLR, low-density lipoprotein receptor; LPL, lipoprotein lipase; PCSK9, proprotein convertase subtilisin/kexin type 9; T2D, type 2 diabetes; TC, total cholesterol; TG, triglyceride; WC, waist circumference.
2.1 Selection of IVs
Independent IVs (r2 < 0.001 and physical distance threshold of 10,000 kb were used to avoid potential linkage disequilibrium [LD]) associated with TG and TC at a genome-wide significance (p < 5 × 10−8) were selected from the Global Lipids Genetics Consortium [8]. Regarding LDL-C, estimates of genetic associations were extracted from summary data of the Genome-wide Association Study (GWAS), including 440,546 individuals in the UK Biobank [9].
On the basis of previous research, the most common drugs used to lower lipid concentrations were chosen. These drugs included 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) inhibitors (statins), proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors, antisense oligonucleotide targeting apolipoprotein B-100 mRNA, and antisense oligonucleotide therapy targeting apolipoprotein C3 (APOC3) mRNA [10, 11] (Table 1). The drug target gene thresholds were identified at a website (https://www.ncbi.nlm.nih.gov/gene). On the basis of the pharmacological effect, the genes targeted by these drugs were classified into two groups. One group of genes comprised those associated with reducing LDL-C concentrations, such as low-density lipoprotein receptor (LDLR), HMGCR, PCSK9, and apolipoprotein B-100 (APOB). The other group of genes comprised those associated with lowering TG concentrations, such as lipoprotein lipase (LPL) and APOC3 (Table 1).
| Primary pharmacological action | Drug class | Substance | Drug targets | Target genes | Gene region(GRCh37/hg19 by Ensembl) | Genetic instruments |
|---|---|---|---|---|---|---|
| Reduced LDL-C | Key regulator | LDL Receptor | LDLR | chr19:11,200,139-11,244,496 | 26 SNPs | |
| HMG-CoA reductase inhibitors | Pravastatin Simvastatin Lovastatin Fluvastatin Atorvastatin Rosuvastatin | HMG-CoA reductase | HMGCR | chr5:74,632,993-74,657,941 | 9 SNPs | |
| Proprotein convertase subtilisin/kexin type 9 inhibitors | Alirocumab Evolocumab | Proprotein convertase subtilisin/kexin type 9 | PCSK9 | chr1:55,505,221-55,530,525 | 19 SNPs | |
| Antisense oligonucleotide targeting ApoB-100 mRNA | Mipomersen | Apolipoprotein B-100 | APOB | chr2:21,224,301-21,266,945 | 12 SNPs | |
| Reduced TG | Key regulator | Lipoprotein Lipase | LPL | chr8:19,796,764-19,824,770 | 8 SNPs | |
| Antisense oligonucleotide targeting ApoC-III mRNA | Volanesorsen | Apolipoprotein C-III | APOC3 | chr11:116,700,623-116,703,788 | 4 SNPs |
- Abbreviations: chr, chromosome; LDL-C, low-density lipoprotein cholesterol; mRNA, messenger ribonucleic acid; SNPs, single-nucleotide polymorphisms; TG, triglyceride.
IVs that proxy for these drug target genes were selected within 100 ± kb of the corresponding genes [11, 12]. IVs with a significance level (p) less than 5 × 10−8 were considered to be strongly associated with the genes targeted by the drug. An LD aggregation threshold (r2 < 0.1) and physical distance threshold (250 kb) were applied to avoid the effect of LD on the results. Within the ±100-kb range of the corresponding gene, a wider LD and physical distance threshold (r2 < 0.1, 250 kb) were applied to capture more genetic variation, including distant loci that may affect the trait or phenotype of interest. This approach also ensured that the variants were more likely to be functionally associated with the drug's target gene and its immediate regulatory regions.
2.2 Data Sources
The primary outcome was osteomyelitis, and this information was obtained in the dataset from the FinnGen consortium (R10). The positive control, coronary heart disease (CHD), was screened from the CARDIoGRAMplusC4D consortium (60,801 cases and 123,504 controls) [13]. All sample information is shown in Supporting Information S1: Table S4. To investigate secondary indicators and mediation effects, we collected datasets (575,531 individuals) containing C-reactive protein (CRP) from genome-wide association study summary statistics [14]. As risk factors for osteomyelitis, data of waist circumference (WC) from the MRC Integrative Epidemiology Unit, type 2 diabetes (T2D) [15], and insulin [16] were selected. The GWAS ID of outcomes, which were identified at a website (https://gwas.mrcieu.ac.uk/), are shown in Supporting Information S1: Table S4.
2.3 Statistical Analysis
Five analytical approaches, namely MR-Egger, weighted median, inverse variance weighted (IVW), simple mode, and weighted mode, were used to assess the effect of lipid traits and drug targets on the outcome. The IVW method was the most frequently used method [12].
Based on the correlation assumption of MR, the F-statistic was used to assess the strength of each genetic variant (an F-statistic > 10 is usually considered to be free of instrumental bias). CHD was chosen as a positive control to verify the validity of the drug target genetic IVs. Furthermore, to support the effect of drug targets on osteomyelitis, CRP (marker of the degree of inflammation and tissue damage) was adopted as a secondary indicator. The relationships between WC, T2D, and insulin with lipid-lowering therapies by genetic agents were evaluated to validate the indirect effects between drug targets and osteomyelitis. If there was a strong association, this implied that there was a potential mediating effect. The MR results were graphically represented in the form of scatter plots (Supporting Information S1: Figure S1).
In contrast to the multivariable MR approach, the two-step cis-MR method was chosen for its ability to mitigate bias arising from high LD correlations among genetic variants in cis-MR analyses. This method offers a distinct advantage in addressing such biases [17].
Sensitivity analyses were conducted using Cochran's Q test, the MR-Egger intercept test, and MR-PRESSO. The MR-Egger intercept test and MR-PRESSO were used to identify horizontal pleiotropy in proxy SNPs, and Cochran's Q value was used to evaluate the heterogeneity of genetic IVs (Supporting Information S1: Table S5). To assess the effect of individual SNPs on the overall outcomes, we used the leave-one-out approach (Supporting Information S1: Figure S2), and systematically removed each SNP and compared the results with those obtained using the IVW method. Bonferroni correction was used to address multiple tests. Our study included three two-sample MR analyses and eighteen drug-target MR analyses. Consequently, the significance thresholds (p) for these two analytical approaches were adjusted to 0.02(0.05/3) and 0.003(0.05/18), respectively. The data analyses were conducted using the TwoSampleMR packages in R version 4.3.1.
3 Results
3.1 Lipid Traits and the Risk of Osteomyelitis
IVs for lipid traits were identified, including 172 independent SNPs associated with LDL-C, 54 associated with TG, and 84 associated with TC (Figure 2, Supporting Information S1: Table S6). We found no association between LDL-C, TG, or TC and osteomyelitis, while sensitivity analysis results for TG and TC suggested the potential presence of horizontal pleiotropy (Supporting Information S1: Table S5). The other results did not show any abnormalities in the heterogeneity test or horizontal pleiotropy test.
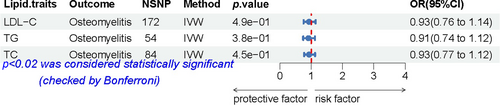
Associations between lipid traits and the risk of osteomyelitis. A forest plot of the association between LDL-C, TG, or TC concentrations and osteomyelitis, as shown by the IVW method. Data are expressed as ORs with 95% CIs (error bars). Associations were considered significant after correcting for multiple testing (0.05/3, p < 0.02). LDL-C, low-density lipoprotein cholesterol; NSNP, number of single-nucleotide polymorphisms; OR, odds ratio; TC, total cholesterol; TG, triglyceride.
3.2 Positive Control Analysis
As anticipated, the findings from the IVW method showed a significant elevation in the risk of CHD associated with PCSK9 (odds ratio [OR] [95% confidence interval [CI]) = 2.09 [1.73–2.53], p = 3.01e−14; Figure 3). Other drug target-positive control results were shown in Supporting Information S1: Table S7. Unfortunately, APOB and LDL-R did not pass the horizontal pleiotropy test in MR-PRESSO. LPL showed horizontal pleiotropy in the MR-Egger intercept test, which indicated that this result lacked robustness (Supporting Information S1: Table S5).
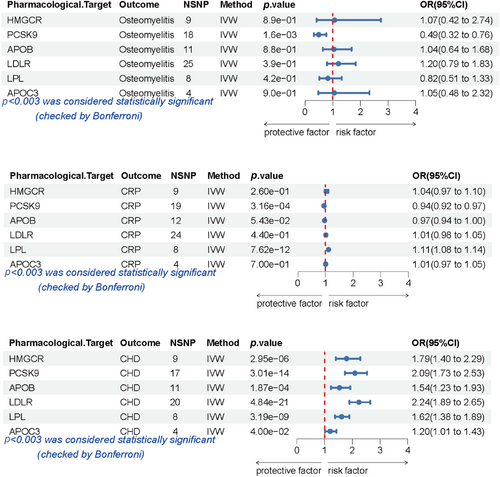
Associations between lipid traits and the risk of osteomyelitis, CHD, and elevated CRP concentrations. A forest plot of the associations between pharmacological targets and CHD, osteomyelitis, or CRP, as shown by the IVW method. Data are expressed as ORs with 95% confidence intervals (error bars). Associations were considered significant after correcting for multiple testing (0.05/18, p < 0.003). APOB, apolipoprotein B-100; APOC3, apolipoprotein C3; CHD, coronary heart disease; CRP, C-reactive protein; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; LDLR, low-density lipoprotein receptor; LPL, lipoprotein lipase; OR, odds ratio; PCSK9, proprotein convertase subtilisin/kexin type 9.
3.3 Lipid-Lowering Drug Targets and the Risk of Osteomyelitis
Nine SNPs that were regarded as genetic IVs were identified in HMGCR, 18 SNPs in PCSK9, 11 SNPs in APOB, 25 SNPs in LDLR, 4 SNPs in APOC3, and 8 SNPs in LPL (Figure 3, Supporting Information S1: Table S7).
Figure 3 shows a genetically proxied association between six lipid-lowering drug classes and their effect on osteomyelitis. PCSK9 was significantly associated with a lower risk of osteomyelitis (OR [95% CI] = 0.49 [0.32–0.76], p = 1.6e−03). Gene mimetics of other drug targets (HMGCR, APOB, LDLR, LPL, and APOC3) showed no significant effect on the prognosis of osteomyelitis. The results of the alternative MR methods were consistent. Moreover, all the results passed the heterogeneity test and the horizontal pleiotropy test (Supporting Information S1: Table S5).
3.4 Lipid-Lowering Drug Targets and CRP Concentrations
The associations between genetic proxies for six lipid-lowering drug targets and CRP concentrations are shown in Figure 3. PCSK9 was associated with lower CRP concentrations (OR [95% Cl] = 0.94 [0.92–0.97], p = 3.16e−04). In contrast, LPL showed adverse effects on CRP (Figure 3, Supporting Information S1: Table S7). Genetic proxy inhibition of drug targets (HMGCR, APOB, LDLR, and APOC3) showed neutral effects on CRP, while the results of LDLR showed heterogeneity (Supporting Information S1: Table S5). With the exception of LDLR, the robustness of all other results was confirmed by the heterogeneity test and the horizontal pleiotropy test (Supporting Information S1: Table S5).
3.5 Mediation Analysis
We conducted a two-step MR analysis to examine the mediating pathway linking PCSK9 to osteomyelitis. We first analyzed the relationship between PCSK9 and WC, followed by an investigation into the association between WC and osteomyelitis. A causal relationship was identified between PCSK9 and WC (p < 0.001, OR = 0.95, 95%CI: 0.92–0.99). A significant association between WC and osteomyelitis was found (p < 0.001, OR = 1.81, 95%CI: 1.41–2.32) (Table 2). These findings suggest that a reduction in the risk of an increase in WC partially mediates the reduced risk of osteomyelitis caused by enhanced PCSK9 levels. However, no association was found between PCSK9 and T2D or insulin. These results were further validated through the heterogeneity test and the horizontal pleiotropy test.
| Exposure | Outcome | Method | nSNP | SE | OR (95%CI) | p |
|---|---|---|---|---|---|---|
| PCSK9 | WC | MR Egger | 15 | 0.02 | 0.96 (0.92–1.01) | 1.25e−01 |
| Weighted median | 15 | 0.02 | 0.96 (0.92–0.99) | 1.22e−02 | ||
| Inverse variance weighted | 15 | 0.02 | 0.95 (0.92–0.98) | 9.17e−04 | ||
| Simple mode | 15 | 0.03 | 0.95 (0.89–1.02) | 1.71e−01 | ||
| Weighted mode | 15 | 0.02 | 0.95 (0.93–0.98) | 1.06e−02 | ||
| WC | Osteomyelitis | MR Egger | 371 | 0.37 | 2.91 (1.42–5.96) | 3.80e−03 |
| Weighted median | 371 | 0.22 | 1.76 (1.15–2.70) | 9.46e−03 | ||
| Inverse variance weighted | 371 | 0.13 | 1.81 (1.41–2.32) | 2.71e−06 | ||
| Simple mode | 371 | 0.66 | 1.79 (0.49–6.58) | 3.80e−01 | ||
| Weighted mode | 371 | 0.41 | 1.93 (0.87–4.28) | 1.06e−01 | ||
| PCSK9 | Type 2 diabetes | MR Egger | 16 | 0.01 | 1.00 (0.99–1.01) | 7.56e−01 |
| Weighted median | 16 | 0.01 | 1.00 (0.99–1.01) | 7.88e−01 | ||
| Inverse variance weighted | 16 | 0.01 | 1.00 (0.99–1.01) | 5.95e−01 | ||
| Simple mode | 16 | 0.01 | 1.01 (0.99–1.04) | 2.24e−01 | ||
| Weighted mode | 16 | 0.01 | 1.00 (0.99–1.01) | 6.95e−01 | ||
| Insulin | MR Egger | 19 | 0.28 | 1.15 (0.66–2.00) | 6.29e−01 | |
| Weighted median | 19 | 0.25 | 1.08 (0.66–1.77) | 7.70e−01 | ||
| Inverse variance weighted | 19 | 0.19 | 0.88 (0.61–1.27) | 4.95e−01 | ||
| Simple mode | 19 | 0.42 | 0.77 (0.34–1.76) | 5.46e−01 | ||
| Weighted mode | 19 | 0.23 | 1.00 (0.64–1.56) | 9.96e−01 |
- Abbreviation: PCSK9, proprotein convertase subtilisin/kexin type 9; SE, standard error; WC, waist circumference.
4 Discussion
This study examined the relationships between osteomyelitis and LDL-C, TC, or TG. Additionally, the associations between six target genes and osteomyelitis were investigated. The target genes were further categorized into LDL-C-lowering targets (PCSK9, HMGCR, APOB, and LDLR) and TG-lowering targets (APOC3 and LPL). Only PCSK9 was associated with a reduced risk of osteomyelitis, providing strong evidence for PCSK9 as a potential therapeutic target for osteomyelitis. A relationship between genetic variants of APOC3 and LPL with lower TG concentrations and osteomyelitis was not found. Additionally, there was a lack of a causal link between TG, APOC3, or LPL and osteomyelitis. The absence of a causal link between HMGCR, APOB, or LDLR and osteomyelitis suggests that PCSK9 reduces the risk of osteomyelitis through alternative mechanisms apart from LDL-C regulation.
PCSK9 is a vital protein involved in the homeostasis of LDL-C and plays a crucial role in the degradation of LDLRs [18]. Some clinical trials support the idea that PCSK9 effectively and safely lowers LDL cholesterol concentrations [19].
PCSK9 may have other effects besides reducing cholesterol. Bone and immune systems are thought to be associated with each other. Some studies have suggested that pro-inflammatory cytokines affect the immune response and bone metabolism. These cytokines enhance macrophage activation, present antigens, and regulate immunity [20].
Previous studies have shown that an increase in PCSK9 during sepsis leads to the activation of the nucleoside binding oligomerization domain-like receptor protein 3 (NLRP3) pathway to induce inflammation [21]. Staphylococcus aureus is an important pathogen in the development of osteomyelitis, driving osteoclast differentiation and enhancing bone resorption. Additionally, the NLRP3 signaling pathway is critically involved in this process [22]. We speculate that PCSK9 activates the NLRP3 signaling pathway because of the critical role of the NLRP3 pathway in the development of sepsis and osteomyelitis. PCSK9 potentially plays a pro-inflammatory role during the pathogenesis of osteomyelitis. CRP is a pivotal and sensitive biomarker for acute-phase responses in the human body, and is frequently used to assess inflammatory activity [23]. Previous studies have acknowledged the role of CRP in diagnosing osteomyelitis [24]. Therefore, we included CRP in a supplementary analysis because significant results could strengthen the association between PCSK9 and osteomyelitis. This association may be caused by exacerbating inflammatory responses. However, further experimental validation is necessary to investigate this possibility.
A clinical correlation between T2D and osteomyelitis has been shown in previous studies. T2D is a risk factor for the development of osteomyelitis. A case report showed that the acute exacerbation of chronic osteomyelitis may be caused by poor blood sugar control in patients with T2D [25]. An MR study showed a negative correlation between PCSK9 expression and the risk of T2D [26]. Although this result was not found through drug target MR analysis in our study, it also partially explains why PCSK9 can inhibit osteomyelitis. Our non-significant result may be attributed to differences in database selection and SNP inclusion/exclusion criteria, necessitating further research to validate this finding.
Using two-step MR, we found that PCSK9 indirectly affected the risk of osteomyelitis by affecting WC. A previous MR study on osteomyelitis reported that an increased WC was positively associated with an increased risk of osteomyelitis, and T2D mediated this relationship [27]. This finding provides insight into the potential role of T2D in the association between PCSK9 and osteomyelitis. WC, as an obesity-related indicator, is widely used to assess the risk of various diseases. In certain specific conditions, WC has greater utility than the body mass index [28, 29]. WC is commonly used to reflect abdominal obesity [30]. Therefore, PCSK9 may affect the risk of osteomyelitis through specific patterns of fat deposition.
Several limitations should be taken into account when interpreting the findings of this study: First, we only analyzed MR in the European population. Therefore, our results may not apply to other populations. Consequently, future investigations should conduct subgroup analyses within diverse populations to attain a more comprehensive understanding. Second, interference with inflammation by bacterial infection may have caused bias. Third, the ability of IV estimation to explain related genetic traits is limited. The mechanisms involved should be determined in further studies.
5 Conclusion
This study shows that there is no relationship between lipid profiles (TG, LDL-C, and TC) and osteomyelitis. We identified several SNPs in PCSK9 as genetic tools to comprehensively assess the influential role of PCSK9 in CHD, CRP, and osteomyelitis. PCSK9 is associated with a lower risk of osteomyelitis, and PCSK9 is also associated with a reduction in CRP concentrations. These findings indicate an association between PCSK9 and osteomyelitis. PCSK9 inhibitors should be used with caution in patients with osteomyelitis or high-risk populations. Our study supports the notion that WC may be a potential mediator between osteomyelitis and PCSK9. Nonetheless, more high-quality studies are necessary to validate our findings, especially large-scale, high-quality prospective studies.
Author Contributions
Zhiyi Zhou: data curation (lead), formal analysis (lead), writing – original draft (lead), writing – review and editing (lead). Zhehan Yang: data curation (lead), formal analysis (lead), methodology (lead), writing – original draft (lead), writing – review and editing (lead). Junpan Chen: data curation (equal), formal analysis (equal), validation (equal), visualization (equal), writing – original draft (equal), writing – review and editing (equal). Minghao Wen: formal analysis (equal), resources (equal), writing – original draft (equal), writing – review and editing (equal). Jiayuan Lei: writing – original draft (equal), writing – review and editing (equal). Wanzhe Liao: writing – original draft (equal), writing – review and editing (equal). Yahan Li: writing – original draft (equal), writing – review and editing (equal). Linghui Liu: writing – original draft (equal), writing – review and editing (equal). Ziyuan Lu: methodology (lead), resources (lead), writing – original draft (lead), writing – review and editing (lead). All authors have granted their approval for the manuscript.
Acknowledgments
We appreciate the time and effort provided by the participants and investigators of the UK Biobank, FinnGen, Global Lipids Genetics Consortium, and the MRC Integrative Epidemiology Unit consortium.
Ethics Statement
No ethics approval was required because all data in the study were previously collected, analyzed, and published.
Consent
The authors have nothing to report.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data shown in the manuscript, code book, and analytical code will be made available upon request pending application.




