Rapid high resolution melting assay to differentiate Streptococcus suis serotypes 2, 1/2, 1, and 14
Graphical Abstract
The designed high resolution melting assay allows to correctly differentiate Streptococcus suis serotype pair 2 and 1/2 or 1 and 14, respectively. The assay is based on a single-nucleotide polymorphism within the capsular polysaccharide synthesis gene cluster K and enables a rapid identification of potential zoonotic serotypes.
Abstract
This rapid high resolution melting (HRM) assay allows distinguishing between Streptococcus suis serotype pairs 2 and 1/2 as well as 1 and 14, respectively, based on a single-nucleotide polymorphism within capsular polysaccharide synthesis gene cluster K. This assay is easy to implement and identifies potential zoonotic serotypes.
1 INTRODUCTION
Streptococcus suis (S. suis) is an important pathogen of pigs and considered to be responsible for various diseases such as septicemia with sudden death, meningitis, endocarditits, and arthritis (Gottschalk, Segura, & Xu, 2007). Currently, there are at least 29 S. suis serotypes (Okura et al., 2016) described based on a serological reaction against the capsular polysaccharide (CPS), which has been described to be a major virulence factor with antiphagocytic properties (Baums & Valentin-Weigand, 2009). S. suis serotype 14 and in particular heterogenous serotype 2 are emerging zoonotic pathogens often associated with disease in both pigs and humans worldwide (Goyette-Desjardins, Auger, Xu, Segura, & Gottschalk, 2014). Formerly, serological typing was performed with different antisera of known type, based on three different techniques: Neufeld's capsular reaction, capillary precipitation, and coagglutination tests (Gottschalk, Higgins, Jacques, & Dubreuil, 1992). In recent times, more frequently multiplex PCR assays are used targeting genes based on CPS (Okura et al., 2014), which are identical for serotype pairs 2 and 1/2, and 1 and 14 except for a single base pair substitution in codon 161 of cpsK gene (Athey et al., 2016). A challenge for diagnostic laboratories is the fact that available PCR tests do not allow resolving these aforementioned serotype pairs 2 and 1/2, and 1 and 14.
Recently, an in silico pipeline using whole-genome sequencing (WGS) short-read data was developed, which is able to differentiate these serotypes (Athey et al., 2016). Nevertheless, for a rapid identification of potential zoonotic strains with serotypes 2 and 14, a reliable inexpensive and feasible high-throughput approach for routine diagnostic laboratories would be a valuable tool. The aim of this study was to evaluate the potential of cpsK as target for differentiating S. suis serotypes 2, 1/2, 1, and 14 from pure culture using a novel high resolution melting (HRM) assay.
2 MATERIALS AND METHODS
For this purpose, four reference strains comprising serotypes 2, 1/2, 1, and 14, one human, and 12 porcine S. suis isolates of serotypes 1 or 14 and 2 or 1/2 were used to develop a novel HRM assay (Table 1). Moreover, 58 S. suis isolates of other serotypes, as well as four further Streptococcus spp. isolates were included in the study. All strains were grown on Columbia agar with sheep blood (Thermo Fisher Diagnostics AG) and incubated at 37°C for 48 hr under aerobic conditions. Strains were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker). Genomic DNA was extracted using a standard heat lysis protocol (Sambrook & Russel, 2006).
| Strains | Strain designation | Source | Year of isolation | Serotypea | Definitive Serotypeb | Sequence typec (ST) |
|---|---|---|---|---|---|---|
| S. suis | ZH 468 | Mitral valve/pigd | 2007 | 2 or 1/2 | 1/2 | ST28 |
| S. suis | ZH 1192 | Lung/pigd | 2015 | 2 or 1/2 | 1/2 | ST28 |
| S. suis | ZH 423 | Heart/pigd | 2016 | 2 or 1/2 | 1/2 | ST1133 |
| S. suis | ZH 1329 | Brain/pigd | 2015 | 2 or 1/2 | 2 | ST1103 |
| S. suis | ZH 462 | Heart/pigd | 2016 | 2 or 1/2 | 2 | ST28 |
| S. suis | ZH 269 | Heart/pigd | 2015 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 1598 | Brain/pigd | 2016 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 1635 | Joint/pigd | 2017 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 1656 | Joint/pigd | 2017 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 730 | Joint/pigd | 2018 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 735 | Joint/pigd | 2018 | 1 or 14 | 1 | ST13 |
| S. suis | ZH 731 | Heart/pigd | 2018 | 1 or 14 | 1 | ST13 |
| S. suis | S. suis14 | Blood/humane | 2018 | 1 or 14 | 14 | ST1 |
| S. suis | Ref. Serotype 1 | Blood/pigf | 1 | 1 | ST13 | |
| S. suis | Ref. Serotype 2 | Brain/pigf | 2 | 2 | ST1 | |
| S. suis | Ref. Serotype 1/2 | Tonsil/pigf | 1/2 | 1/2 | ST56 | |
| S. suis | Ref. Serotype 14 | Not known/humanf | 14 | 14 | ST6 | |
| 58 S. suis | — | Various tissues/pigd | 2007–2017 | 13 different serotypesg | — | 29 different ST |
| 4 S. spp. | — | Various tissues/pigd | 2007–2012 | — | — | — |
- a Serotype characterization by multiplex PCR (Kerdsin et al., 2014).
- b Definitive serotype assignment after S. suis HRM.
- c Sequence type characterization by multilocus sequence typing (King et al., 2002).
- d Strains isolated between 2007 and 2018 from the Department of Veterinary Bacteriology, University of Zurich, Switzerland.
- e Bacteriology Laboratory, Division of Laboratory Medicine, Geneva University Hospitals, Geneva, Switzerland.
- f Reference strains from Swine and Poultry Infectious Diseases Research Center, Groupe de recherche sur les maladies infectieuses des animaux de production, University of Montreal, Saint-Hyacinthe, Canada.
- g Serotypes 3, 4, 5, 6, 7, 8, 9, 12, 15, 16, 21, 28, and 31 (belonging to 29 different sequence types) were tested.
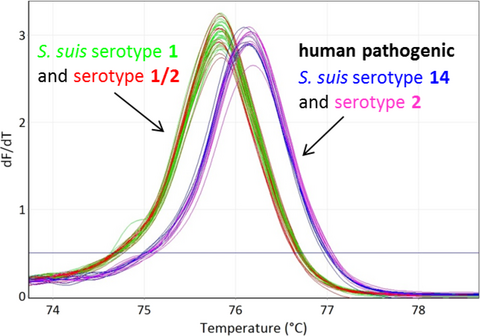
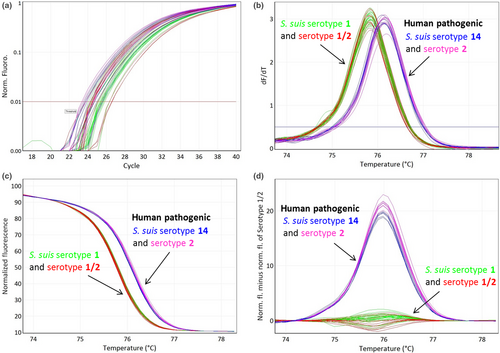
The cpsK region flanking the nonsynonymous single-nucleotide polymorphism (SNP) at base pair position 483 (Athey et al., 2016; Roy et al., 2017) (G for serotypes 2 and 14, T or C for serotypes 1 and 1/2) was targeted using the following primers: (cpsK_for: 5′- GATGGTCATCGCTTTGTGGTG-3′) and (cpsK_rev: 5′- GAGCAAGCGATAAGTGAAGTATTCATC-3′) and producing an amplicon of 117 bp (Figure 1). All qPCR experiments with subsequent HRM analysis were performed on a Rotor-Gene Q (Qiagen, Hilden, Germany) using Type-it HRM PCR Kit (Qiagen). The total reaction volume was 15 µl. About 1 µl of sample DNA of 100 pg was added to a reaction mixture containing 7.5 µl 2× Type-it HRM Mastermix, 0.5 µM of each primer and ultrapure water. The PCR thermocycling conditions were as follows: initial denaturation at 95°C for 5 min, 40 cycles with denaturation at 95°C for 10 s, and annealing/extension at 66°C for 30 s followed by a second cycling step at 95°C for 10 s and 40°C for 2 min. Finally, a HRM ramping from 70°C to 82°C was performed. Fluorescence data were acquired at 0.1°C increments every 2 s to generate specific melting curves. Reference strains with serotypes 2, 1/2, 1, and 14 were included as melting curve standards and positive controls. To exclude contaminations in the reaction mixture, ultrapure water was used as a negative control in each experiment. Data analysis was performed using Rotor-Gene Q Software 2.3.1 (Qiagen). Normalized and difference plots were generated. To normalize the results, the premelt and postmelt signals of all samples were set to uniform relative values from 100% to 0%. In order to generate difference plots, normalized fluorescence data of sample curves were subtracted from the curve of reference strain serotype 1/2 to visually accentuate differences in a greater resolution.

The HRM assay was evaluated, and its specificity was determined. To examine the intra- and interassay variability of the melting temperatures (Tm), representing the repeatability of the developed HRM assay, all isolates were tested. The variability assays were performed in triplicates in three independent runs at three different days.
3 RESULTS
The HRM assay clearly divided the 17 S. suis strains into two clusters grouping serotype 2 with 14 and serotype 1/2 with 1 (Figure 2). By combining serotype determination obtained by PCR (Kerdsin et al., 2014) into serotype pairs 2 and 1/2 or serotype pairs 1 and 14 with the obtained results of the HRM assay, it is possible to separate and correctly assign the corresponding serotype to each S. suis isolate. Based on our results, the target was 100% specific for the four serotypes tested as none of the Streptococcus spp. or S. suis with different serotypes (Table 1) yielded a PCR positive result. The intra-assay coefficients of variability (CVs) of Tm were between 0.008% and 0.03% and interassay CVs, respectively, between 0.04% and 0.07% illustrating a highly reproducible and stable assay. Serotypes 1 and 1/2 yielded a Tm of 75.9 ± 0.1°C, whereas serotypes 2 and 14 yielded a Tm of 76.2 ± 0.1°C, respectively (Figure 1).
4 DISCUSSION
In this report, a novel HRM assay based on SNP detection in PCR products is described. HRM analysis is a rapid and low cost genotyping method simply convertible in laboratories as a singleplex method with a low risk of contamination (Reed, Kent, & Wittwer, 2007). The reliable distinction between serotypes 2, 14, 1, and 1/2 is useful for classification of circulating S. suis strains and helps strain tracking in case of disease outbreaks. In addition, this assay provides an easily applicable diagnostic tool for high-throughput screening and rapid identification of S. suis serotypes 2 and 14 relevant for human infection without the need of a WGS approach. Compared with Sanger sequencing and WGS, a HRM assay is not depended on manual inspection of the sequencing data and results are obtained straightforward in less than 1.5 hr. Reagent costs are low with about 1 CHF per sample in comparison to Sanger sequencing, which was around ten times as expensive with an estimated prize of 10 CHF per sample. The cost of WGS sequencing of approximately 200 CHF per sample is not competitive if the focus of investigation is set at serotype differentiation or at screening of potentially zoonotic isolates. Even though, the local reagents and labor cost may be variable, HRM is by far the most cost efficient and easily applicable assay delivering accurate results in a short turnaround time. Thus, a possible surveillance system contributing to the development of proficient public health policies can be established.
5 CONCLUSION
To conclude, we have developed a specific HRM assay distinguishing between serotype pairs 2 and 1/2 and 1 and 14, respectively, based on a SNP within cpsK. This assay allows a rapid separation of these S. suis serotypes in routine diagnostic laboratories using molecular based serotyping in order to assess a zoonotic risk of emerging strains.
ACKNOWLEDGMENTS
The authors wish to thank the laboratory staff of the Department of Veterinary Bacteriology, Vetsuisse Faculty, University of Zurich, for excellent technical assistance.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTIONS
Simone Scherrer: Conceptualization-Equal, Investigation-Equal, Validation-Lead, Writing-original draft-Lead; Fenja Rademacher: Investigation-Supporting; Nathalie Spoerry Serrano: Investigation-Supporting; Jacques Schrenzel: Resources-Equal, Writing-review and editing-Supporting; Marcelo Gottschalk: Resources-Equal, Writing-review and editing-Supporting; Roger Stephan: Conceptualization-Equal, Supervision-Lead, Writing-review and editing-Lead; Patricia Landolt: Conceptualization-Equal, Investigation-Equal, Validation-Supporting, Writing-review and editing-Supporting.
ETHICS STATEMENT
None required.
Open Research
DATA AVAILABILITY STATEMENT
Raw datasets from intra- and interassay variability runs are available upon request.