Recombinant spider silk coatings functionalized with enzymes targeting bacteria and biofilms
Graphical Abstract
Surgical implants could potentially be protected from colonization of bacteria by having a protective coating with antibacterial properties. Herein, we investigate strategies to use assembly of silk functionalized with antibacterial enzymes to prepare such coatings. Both the peptidoglycan degrading enzyme SAL-1 and the biofilm-matrix degrading enzyme Dispersin B were shown effective when immobilized using a silk coating.
Abstract
Bacteria forming biofilms on surgical implants is a problem that might be alleviated by the use of antibacterial coatings. In this article, recombinant spider silk was functionalized with the peptidoglycan degrading endolysin SAL-1 from the staphylococcal bacteriophage SAP-1 and the biofilm-matrix-degrading enzyme Dispersin B from Aggregatibacter actinomycetemcomitans using direct genetic fusion and/or covalent protein–protein fusion catalyzed by Sortase A. Spider silk assembly and enzyme immobilization was monitored using quartz crystal microbalance analysis. Enzyme activity was investigated both with a biochemical assay using cleavage of fluorescent substrate analogues and bacterial assays for biofilm degradation and turbidity reduction. Spider silk coatings functionalized with SAL-1 and Disperin B were found to exhibit bacteriolytic effect and inhibit biofilm formation, respectively. The strategy to immobilize antibacterial enzymes to spider silk presented herein show potential to be used as surface coatings of surgical implants and other medical equipment to avoid bacterial colonization.
1 INTRODUCTION
Bacteria, including pathogens such as Staphylococcus aureus, can attach to and grow biofilms on almost any surface (Hall-Stoodley, Costerton, & Stoodley, 2004), which thus occasionally occur on surgical implants and at surgical sites (Swartjes et al., 2013). A biofilm has a three-dimensional extracellular structure consisting of polysaccharides, proteins, nucleic acids, and lipids, which are secreted by the bacteria, and varies between different species (Abee, Kovács, Kuipers, & Veen, 2011; Karatan & Watnick, 2009). The biofilm protects the bacteria from antibiotics by providing a physical barrier hindering the antibiotic to reach them. Dormant bacteria usually reside within the biofilm, which furthers their resistance against antibiotics (Abee et al., 2011).
In order to decrease the risk of infections related to surgery, many research groups have worked on creating antibacterial coatings protecting against various bacterial strains. There are several methods based on different active substances, which have been tried with varying success. Coatings created using polycationic polymers, chitosan and alginates have been reported to give an antibacterial effect (Gomes, Mano, Queiroz, & Gouveia, 2013). Also, coatings containing N-halamines (Gomes et al., 2013) and silver ions as active substances have been shown effective (Umair et al., 2016). PEG-coating has been proven capable of preventing biofilm formation by inhibiting the binding of proteins which often precede bacterial attachment (Chen, Li, Zhao, & Zheng, 2010). To achieve a local antibacterial activity, it is favorable to immobilize an enzyme on the surface. This can be done for example by chemical or enzymatic conjugation. Swartjes et al. (2013) created a polymethylmethacrylate coating onto which DNase-1 was attached via dopamine, which inhibited biofilm formation. Silk can be used as another example of coating suitable for in vivo usage, since it is biocompatible and nontoxic (Widhe et al., 2010). The recombinant spider silk protein 4RepCT has previously been fused to various fold-dependent bioactive domains, which showed retained activity after assembly into fibers or coatings (Jansson et al., 2014; Nilebäck, Chouhan, et al., 2017; Nilebäck, Hedin, et al., 2017). It is thus a promising base for a surface coating to present domains that prevent and disrupt biofilms. In this article, the enzyme Dispersin B (DspB) and the enzymatic domain SAL-1 was attached to 4RepCT in an attempt to create antibacterial and biofilm-preventing coatings. DspB is a biofilm-matrix-degrading enzyme produced by the bacteria Aggregatibacter actinomycetemcomitans to degrade its own biofilm during active spreading. DspB targets poly-β-1,6-N-acetyl-d-glucosamine, which is found also in the biofilms of, for example, S. aureus (Tews et al., 1996). SAL-1 is an endolysin from the bacteriophage SAP-1, which break bonds in the peptidoglycan in order to degrade the cell wall of the host bacteria.
We present two approaches to functionalize silk coatings with these two enzymes; functionalization before assembly, preassembly and functionalization after assembly, postassembly. In the preassembly approach, the enzyme and silk protein are expressed recombinantly as a fusion protein. The enzyme-silk fusion protein then self-assembles into silk coatings via intermolecular interactions between the silk parts. In the postassembly approach, the silk proteins and the enzyme are produced separately. Coatings are first made by self-assembly of the silk protein, and the enzyme, which has been designed to include a short peptide recognition motif for a transpeptidase at its C-terminus, is then conjugated to the silk coating by the use of the transpeptidase Sortase A. Both the preassembly and postassembly approaches were used in this project, to explore their possibilities and limitations. Immobilization of the enzymes was evaluated using quartz crystal microbalance (QCM) analysis. The catalytic ability of DspB and SAL-1 after immobilization onto silk coatings was assessed in biochemical assays using substrate analogues. Finally, their ability to inhibit biofilm and lyse bacteria, respectively, was examined in bacterial assays.
2 MATERIALS AND METHODS
2.1 Chemicals and reagents
All chemicals and reagents were bought from Sigma-Aldrich if not stated otherwise.
2.2 Protein expression and purification
The silk protein 4RepCT was generously provided by Spiber Technologies AB in 20 mM tris(hydroxymethyl)aminomethane (Tris) buffer, pH 8.0, and stored frozen. A variant of Sortase A with three mutations (P94S, D160N, K196T) introduced to enhance its catalytic activity was used and is hereafter denoted Sortase A with the mutations (SrtA*). The vector was kindly provided by Dr. Kristina Westerlund and used to produce SrtA* as previously described (Altai et al., 2017).
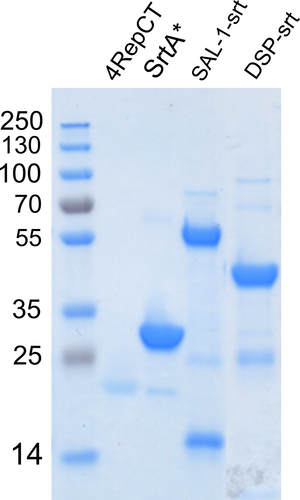
The sequence for DspB was taken from PDB-ID: 1YHT and the sequence for SAL-1 was derived from the GenBank sequence AFN38718.1, although with the amino acid changes V26I, E113Q, and Q485H (*) described by Jun et al. (2017). DspB and SAL-1 was adapted to SrtA-mediated conjugation by adding a C-terminal tag at the end including glycine/serine-rich linkers and the SrtA recognition tag (srt), that is, the peptide sequence LPETG as described previously (Nilebäck et al., 2019), resulting in the following sequence design: GSSG-H6-1YHT-GSG-PAAL-GSAS-(GS)3-LPETGG, denoted DspB with a sortase recognition tag (DspB-srt) and AFN38718.13*-GSSG-PAAL-GSSG-LPETG-H6, denoted SAL-1-srt.
Dispersin B with a sortase recognition tag was produced recombinantly in Escherichia coli BL21* (DE3) by cultivation at 150 rpm, 37°C and induction of expression by addition of 0.5–1 mM isopropyl-β-d-thiogalactopyranoside. Cultivation was continued at 25°C overnight. Harvesting was done by centrifugation, and the pellet was resuspended in 20 mM Tris/200 mM NaCl, pH 8.0. A similar protocol was used for expression of SAL-1-srt, with overnight expression at ambient temperature, and the pellet was dissolved in 50 mM Tris/300 mM NaCl/30% glycerol/10 mM imidazole, pH 8.0. Prior to purification, the cell suspensions were thawed, lysed by sonication and centrifuged. The supernatant of SAL-1-srt was further incubated for 30 min with 8.8 U/ml DNAseI and 1.7 µl/ml 1 M MgCl2 to reduce the viscosity. The SAL-1-srt suspension was incubated with Ni-NTA resin (Qiagen, Venlo, Netherlands) and rotated at 4°C for 2 hr before packing into columns. After wash, elution was done with 50 mM Tris/300 mM NaCl/30% glycerol/250 mM imidazole. DspB-srt was added to a prepacked Ni-NTA column, washed before elution in 20 mM Tris/200 mM NaCl/200 mM imidazole. Purity was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
The DspB-4RepCT silk fusion protein was recombinantly expressed from a pT7-based vector providing a C-terminal GSSG-His₆tag and purified essentially as described above (SAL-1-srt) and thereafter dialyzed toward phosphate-buffered saline (PBS). DspB-4RepCT was stored frozen at −20°C.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of the purified proteins is shown in Figure A1.
2.3 Surface adsorption analysis
Quartz crystal microbalance with Dissipation (QCM-D) was made as described previously (Nilebäck, Hedin, et al., 2017). Equilibration was done with 20 mM Tris. About 0.1 g/L of either 4RepCT or DspB-4RepCT in 20 mM Tris was flowed over the sensor at 20 µl/min for 120 min, and the surfaces were then rinsed with 20 mM Tris.
2.4 Quartz crystal microbalance analysis of Sortase A coupling of SAL-1-srt and DspB-srt to silk coating
Polystyrene sensor chips (untreated, PS 0292645) were coated by incubation of 200 µl of 0.1 mg/ml 4RepCT silk protein solution (20 mM Tris) onto the sensor surface for 1 hr. This was followed by three times wash with 20 mM Tris. The coupling analysis was then performed using Attana Cell 200 (Attana AB). The flow rate was set to 1 µl/min, and the injection volume for each injection was 35 µl. The reaction buffer contained 4.3 µM DspB-srt or SAL-1-srt, and 3 µM SrtA* prepared in ligation buffer (50 mM Tris, 150 mM NaCl, 10 mM CaCl2, pH 7.5). Sample injections lasted 35 min and were followed by 20 min' re-establishment of baseline with running buffer (reaction buffer excluding DspB-srt or SAL-1-srt and SrtA). Blank injections were done with the running buffer.
2.5 Coupling of enzymes SAL-1-srt and DspB-srt to silk 4RepCT
Silk coatings for preassembly were prepared by incubation of 200 µl of 4.3 µM 4RepCT solution in 96-well suspension plates (Sarstedt). After 30 min, the solution was removed and the coatings were washed three times with 20 mM Tris. The coatings were then top-coated with 1.5 µM (0.1 g/L) of DspB-4RepCT for 30 min and rinsed three times with 20 mM Tris. The coatings were kept in buffer until the start of experiments. Silk coatings for postassembly functionalization were prepared in the same way, by incubation of 200 µl of 4.3 µM 4RepCT solution in 96-well suspension plates for 1 hr, followed by three times wash with 20 mM Tris. For SrtA*-mediated conjugation, 200 µl reaction mixture was added to each well, including 3 µM SrtA*, 4.3 µM DspB-srt or SAL-1-srt in ligation buffer. After 2 hr incubation at ambient temperature, the wells were washed three times with 200 µl PBS. Negative controls were either untreated silk coatings or silk coatings incubated with reaction mixtures containing SrtA* storage buffer instead of SrtA*, or ligation buffer instead of DspB-srt/SAL-1-srt.
2.6 Biochemical 4-nitrophenyl N-acetyl-β-d-glucosaminide assay for DspB
A rate assay method with 4-Nitrophenyl N-acetyl-β-d-glucosaminide (NP-GlcNAc; Sigma-Aldrich) was used to determine the enzyme activity of DspB coupled to 4RepCT using SrtA*.
Samples were prepared using the pre- or postassembly method, after which the samples were washed with 50 mM phosphate buffer, pH 7.0. About 100 µl of 2 mg/ml NP-GlcNAc in 50 mM phosphate buffer was added to the wells and kept at ambient temperature. The reaction was terminated after 72 hr by the addition of 10 µl 1M NaOH. The absorbance at 405 nm was measured using a ClarioStar plate reader (BMG Labtech).
2.7 Biochemical fluorescein Di-β-d-galactopyranoside assay for SAL-1
In the presence of SAL-1, the substrate Fluorescein Di-β-d-Galactopyranoside (FDG) is degraded to fluorescein that can be detected using fluorescence. Samples were prepared using 100 µl sample volumes in 96-well plates. After SrtA*-mediated conjugation, the samples were washed with PBS. The plate was first run in a plate reader for 120 cycles, 60 s cycle time, 40 flashes/well without addition of FDG substrate to simulate the possible wash effects that could lead to removal of nonspecifically surface-bound SAL-1-srt. The supernatants were then moved from one of the lanes to remove any SAL-1-srt that had been washed into the solution, and leave SAL-1-srt that was still surface-bound in the well. About 100 µl of PBS was added to these wells so that all wells had the same amount of liquid. Then, 100 µl of 10 µg/ml FDG (Invitrogen) in PBS was added to each well and the program was run again, with λexcitation = 490 nm and λemission = 525 nm, measured every 60 s during 120 cycles.
2.8 Quantitative biofilm assay with crystal violet
Coupling of 4RepCT and DspB-srt was done as described above. Staphylococcus aureus was grown overnight, washed and resuspended in PBS to OD600 = 0.8. About 200 µl of suspended bacteria was added to each well, and the plate was incubated at 37°C without shaking for 24 hr. After the incubation, the liquid was removed and the wells were washed two times with 200 µl PBS. For fixation, 100 µl 95% ethanol was added to each well and the plate was incubated for 10 min at ambient temperature without shaking, and then washed three times with 100 µl MilliQ water. The wells were stained with 50 µl 0.1% Crystal Violet at ambient temperature without shaking for 30 min. The Crystal Violet solution was removed and the wells were washed 4 times with 100 µl MilliQ water. About 40 µl of HAc/Acetic Acid was added to each well, and the plate was incubated at 200 rpm at ambient temperature for 20 min. About 35 µl of the acetic acid was transferred from each well to new wells. The samples were measured at 595 nm in a Polar Star Omega plate reader (BMG Labtech) at discrete wavelength and a settling time of 0.5, it was shaken at 500 rpm in double orbital before measurement.
2.9 Turbidity reduction assay with SAL-1-srt
TSB broth (Oxoid; Thermo Fisher Scientific) was incubated with overnight culture of S. aureus[SA113] and grown to OD600 = 0,4. The cells were washed and resuspended in cold 10 mM Phosphate/150 mM NaCl/25% glycerol, pH 7.5, and stored at −80°C.
SAL-1-srt was coupled to 4RepCT as described above. Substrate cells were added to a total volume of 100 µl with OD600 = 1. The absorbance was measured in a Polar Star Omega at 595 nm. Measurements were made once per minute, while shaking the plate at 200 rpm in “orbital shaking”-mode before each measurement cycle and at 200 rpm in “meander corner shaking”-mode between measurement cycles.
2.10 Statistical analysis
Standard deviations were calculated for sample replicates within an experiment, and error propagation was performed for experimental replicates to obtain a corrected standard deviation per sample type. One-way analysis of variance (ANOVA) was conducted in GraphPad Prism with following multiple comparisons of means using Tukey's test.
3 RESULTS AND DISCUSSION
3.1 Strategies for immobilization of enzymes onto coatings of recombinant spider silk
The partial spider silk protein 4RepCT has previously been reported to readily self-assemble into a stable fibrillar coating on solid surfaces. This is seen by a characteristic continuous build-up of proteins on the surface as long as proteins are available in the bulk solution. A continuous build-up of assembling proteins has been observed also for 4RepCT silk proteins produced in fusion with short peptide motifs (Nilebäck, Hedin, et al., 2017). This behavior is different compared with surface adsorption of most other proteins, which instead of a continuous build-up stagnates after filling the surface (Horbett, 2004).
Herein, the biofilm-preventing enzyme DspB was produced in fusion with 4RepCT, in order to investigate the possibilities to use the preassembly approach to prepare silk coatings with the capacity to inhibit biofilm formation. First, the ability of the DspB-4RepCT fusion protein to self-assemble on solid surfaces was assessed using QCM measurements. However, compared with the surface adsorption of 4RepCT, the fusion protein DspB-4RepCT does not show a characteristic silk assembly phase (Figure 1a). Likely, sterically hindrance by the added domain restricts the silk part from interacting with other silk proteins. In a previous study (Thatikonda, Nilebäck, Kempe, Widhe, & Hedhammar, 2018), similar adsorption pattern was observed for 4RepCT in fusion with another folded domain; the basic fibroblast growth factor (FGF2). In order to obtain stable FGF2-silk coatings, a thin coating of 4RepCT was therefore first prepared, which was then coated with FGF2-silk on top. Herein, the same two-step coating strategy was used to obtain a sufficient surface coverage of silk fused with catalytically active DspB.
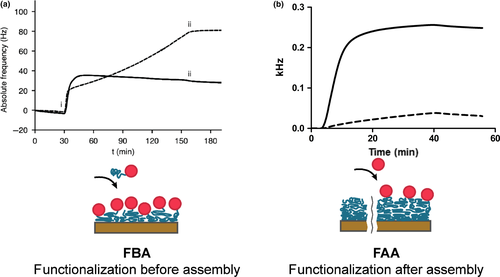
As alternative, a postassembly approach to prepare silk coatings that are functionalized with DspB after assembly was also investigated. First, 4RepCT silk proteins were allowed to assemble into a coating on a polystyrene surface. Then, a mixture of DspB-srt and SrtA* was added at a slow flow, for a contact time of 35 min. Thereby, DspB-srt is covalently coupled to the N-terminal glycine on 4RepCT in the silk coating, by the action of SrtA*. Successful immobilization of DspB onto the silk coating was observed as a frequency change corresponding to mass adsorption (Figure 1b).
3.2 Catalytic ability of DspB immobilized on silk coatings
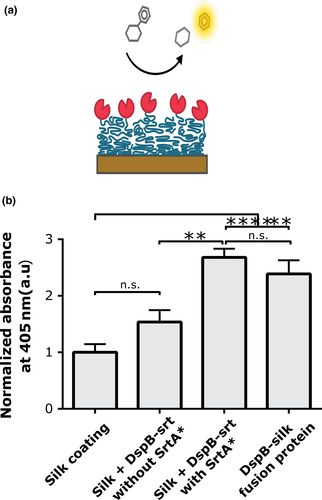
The catalytic ability of DspB immobilized on silk coatings by either the pre- or postassembly strategy was first assessed using a colorimetric assay with a substrate analogue.
NP-GlcNAc is a substrate analogue to the polymer in biofilms (GlcNAc) that is degraded by DspB, modified to release nitrophenol when the glycosidic linkage in the substrate is cleaved. An increase in absorbance at 405 nm, due to the presence of released nitrophenol, was measured from all samples incubated on silk coating with DspB, compared to the control with bare silk coatings (Figure 2). The highest signals were obtained from coatings prepared using the postassembly approach, when DspB-srt was added with SrtA* to silk coatings. A high activity was also obtained from silk coatings prepared using the preassembly approach using the DspB-4RepCT fusion protein. A slight increase in absorbance was observed for controls where DspB-srt had been incubated on the silk coatings without SrtA*, although not statistically significant compared to the controls with bare silk.
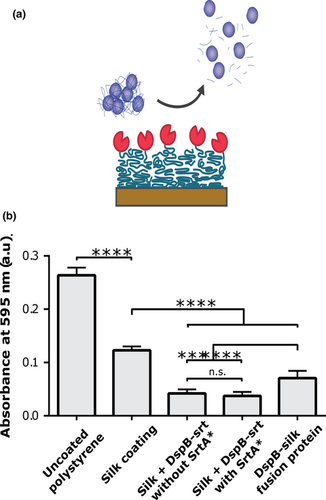
3.3 Biofilm formation on silk coatings
Next, the capability of DspB to reduce the bacterial load in a biofilm assay after immobilization onto silk coatings was evaluated. Bacteria were cultivated on the different coatings during 24 hr, and the total bacterial load was subsequently quantified using Crystal Violet staining (Figure 3). The highest values were obtained for bacteria cultured on bare plastic wells. A lower amount of bacteria grew on coatings of only silk than the bare plastic. However, significantly lower bacteria load was obtained on silk coatings functionalized with DspB, prepared either using the pre- or postassembly approach. Lowest bacteria load was obtained using DspB-srt conjugated to silk in presence of SrtA*. Also in this assay, silk coatings where DspB-srt had been incubated without SrtA* gave a measurable effect.
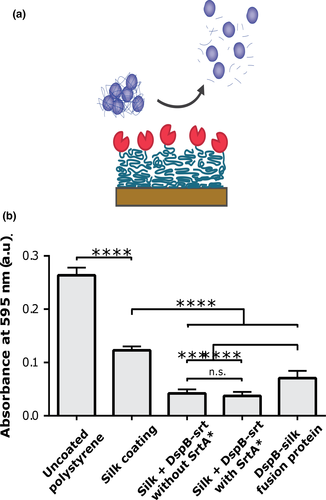
3.4 Immobilization of a two-domain enzyme to silk coatings
Encouraged by the observed activity of silk coatings functionalized with DspB, we decided to investigate the possibility to use a similar approach also for an endolysin. SAL-1 is an endolysin that has previously been shown effective to cleave peptidoglycans to degrade the cell wall of bacteria. The enzyme is composed of two domains, and therefore a bit more demanding for a recombinant expression system. Moreover, fusion of silk with two domains would likely limit the assembly ability of the silk part even more. This, and the similarities in results obtained from pre- and postassembly of DspB, motivated us to only utilize the postassembly approach in this case.
Quartz crystal microbalance analysis showed that SAL-1 with a sortase recognition tag (SAL-1-srt) was successfully coupled to a silk coating when added together with SrtA* at a slow flow (Figure A2).
3.5 Catalytic ability of SAL-1 when immobilized on silk coatings
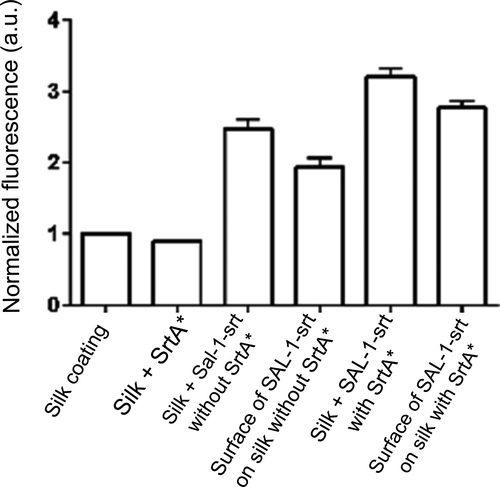
Recombinantly produced CHAP domain from the SAL-1 homolog LysK has previously been found able to convert the substrate Fluorescein Di-β-d-Galactopyranoside (FDG) to a fluorescent product (Seijsing J and Jönsson HN, unpublished). It can be assumed that the CHAP domain in SAL-1 targets the same bond as the CHAP domain in LysK, being the bond connecting the d-Alanine and Penta-Glycine (Jun et al., 2011). This was herein utilized to assess if SAL-1 retains its catalytic ability after conjugation to silk coatings. FDG solutions incubated on silk coatings with SAL-1-srt showed significantly higher levels of fluorescence compared with those incubated on bare silk coating (Figure 4). However, FDG solutions incubated on silk coatings prepared with SAL-1-srt in absence of SrtA* also showed some increased fluorescence, indicating the occurrence of unspecific binding of SAL-1-srt to the silk coatings. All silk coatings were thoroughly washed (three times) before the assessment of their catalytic ability using FDG, in order to measure the effect of surface-bound SAL-1-srt rather than released in the bulk. Moreover, the coatings were first incubated in just buffer, to release any unspecific bound SAL-1-srt. By addition of FDG substrate to this incubation buffer, it was concluded that the main portion of SAL-1-srt, whether StrA was included or not, remained on the surface throughout the experiment rather than being released to the buffer (Figure A3).
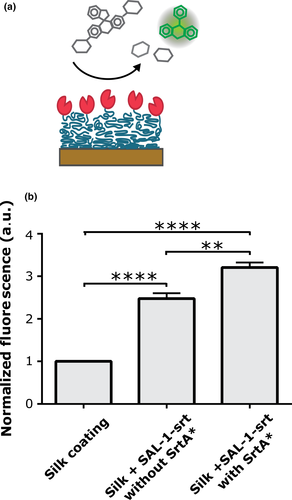
3.6 Bacteriolytic ability of silk coatings
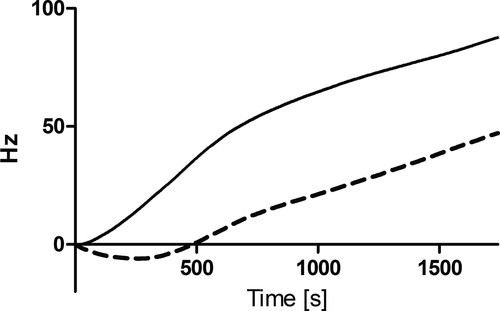
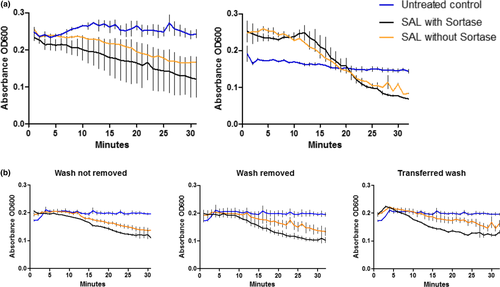
The ability of SAL-1-srt to lyse bacteria was investigated using a turbidity reduction assay (TRA) with S. aureus in suspension. Upon lysis of bacteria, a decrease in optical density of the bacteria suspension is observed. However, this assay requires that the bacteria in suspension come in contact with the coating where SAL-1-srt is immobilized, why low reduction rates are expected. A decrease in absorbance, corresponding to the amount of lysed bacteria, was seen for suspensions incubated on silk coatings with SAL-1-srt, compared with bare polystyrene wells (Example in Figure 5, replicates in Figure A4). Although the reduction rates varied between experiments, the absorbance were decreased more for all bacteria suspensions incubated on silk coatings prepared with SAL-1-srt and SrtA* compared with the controls in all experiments.
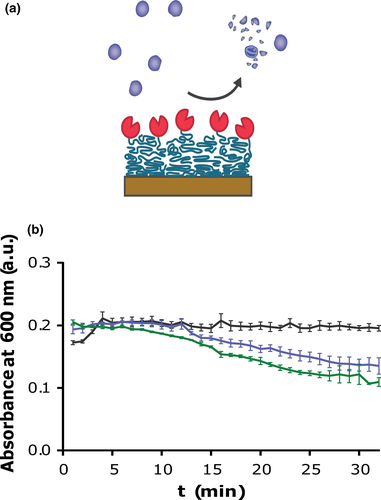
4 CONCLUSIONS
In this work, we evaluated two approaches to immobilize two different enzymes that target bacteria onto silk coatings. The preassembly strategy allows a high level of control over the covalent attachment but a low flexibility for expression and low ability to predict how the functional domain influences the silk assembly. The postassembly approach is easy to conduct, adapt, and optimize, although the conjugation efficiency is a limiting factor. Both methods show potential for construction of bioactive surfaces with antibacterial properties or antibiofilm properties.
ACKNOWLEDGMENTS
We thank Kristina Westerlund (Royal Institutes of Sweden) for providing the Sortase A* construct and for guidance on its properties and use. We thank Spiber Technologies AB for providing silk proteins. This work was funded by Formas (to MH), Swedish Research Council (to MH), Knut and Alice Wallenbergs stiftelse (to MH), the Sven and Lilly Lawski Foundation (to JS); the Olle Engkvist Byggmästare Foundation [2015/419] (to Anders S. Nilsson and JS) and the Skolar Award (to JS).
CONFLICT OF INTERESTS
M.H. holds shares in Spider Technologies AB, which aims to commercialize recombinant spider silk.
AUTHOR CONTRIBUTIONS
Fredrik Seijsing: Formal analysis; Investigation; Writing-original draft. Linnea Nilebäck: Data curation; Formal analysis; Investigation; Supervision; Writing-original draft. Oskar Öhman: Formal analysis; Investigation. Rajeev Pasupuleti: Formal analysis; Investigation. Camilla Ståhl: Formal analysis; Investigation. Johan Seijsing: Conceptualization; Project administration; Supervision; Writing-review & editing. My Hedhammar: Conceptualization; Project administration; Supervision; Writing-review & editing.
ETHICS STATEMENT
None required.
Appendix 1