The metagenomic landscape of a high-altitude geothermal spring in Tajikistan reveals a novel Desulfurococcaceae member, Zestomicrobium tamdykulense gen. nov., sp. nov
Graphical Abstract
Abstract
Metagenomic analysis was conducted to assess the microbial community in the high-altitude Tamdykul geothermal spring in Tajikistan. This analysis yielded six high-quality bins from the members of Thermaceae, Aquificaceae, and Halothiobacillaceae, with a 41.2%, 19.7%, and 18.1% share in the total metagenome, respectively. Minor components included Schleiferia thermophila (1.6%) and members of the archaeal taxa Pyrobaculum (1.2%) and Desulfurococcaceae (0.7%). Further analysis of the metagenome-assembled genome (MAG) from the Desulfurococcaceae family (MAG002) revealed novel taxonomy with only 80.95% closest placement average nucleotide identity value to its most closely related member of the Desulfurococcaceae family, which is part of the Thermoproteota phylum comprising hyperthermophilic members widespread in geothermal environments. MAG002 consisted of 1.3 Mbp, distributed into 48 contigs with 1504 predicted coding sequences, had an average GC content of 41.3%, a completeness and contamination rate of 98.7% and 2.6%, respectively, and branched phylogenetically between the Ignisphaera and Zestosphaera lineages. Digital DNA-DNA hybridization values compared with Ignisphaera aggregans and Zestosphaera tikiterensis were 33.7% and 19.4%, respectively, suggesting that this MAG represented a novel species and genus. Its 16S rRNA gene contained a large 421 bp intron. It encodes a complete gluconeogenesis pathway involving a bifunctional fructose-1,6-bisphosphate phosphatase/aldolase; however, the glycolysis pathway is incomplete. The ribulose monophosphate pathway enzymes could be used for pentose synthesis. MAG002 encodes several hydrogen-evolving hydrogenases, with possible roles as hydrogen sinks during fermentation. We propose the name Zestomicrobium tamdykulense gen. nov. sp. nov. for this organism; it is the first thermophilic genome reported from Tajikistan.
1 INTRODUCTION
Extreme environments, such as hot springs, submarine hot vents, salt lakes, soda lakes, and volcanic regions, represent a rich source of extremophilic bacteria and archaea with great potential for use in industrial and biotechnological processes (Egamberdieva et al., 2018; Kuddus, 2021). Thermophilic microbes growing at 50 to 100°C provide many thermostable hydrolytic enzymes such as proteases, lipases, amylases, cellulases, etc. which are exploited in industrial and biotechnological processes globally (Atalah et al., 2019; Kuddus, 2021; Satyanarayana et al., 2013). Terrestrial thermal springs are widely distributed worldwide and vary greatly in temperature, pH, and redox level, which are factors that drive evolution and shape the diversity and community structure of hot spring microbiota (Inskeep, 2013). Thermal springs are classified according to the origin of the water: magmatic water originating in volcanic areas and hot telluric water formed when groundwater passes along deep hot rocks (Arsanova, 2014), which further affects water mineralization and ultimately drives the evolution and diversity of microbial communities (Castelán-Sánchez et al., 2020).
During the last decade, metagenomics-based assessments of geothermal environments have boosted our understanding of microbial community structures in these habitats and the mining of novel thermostable enzymes of industrial and biotechnological potential (Chopra et al., 2020; Kuddus, 2021; Kumar, 2023; Nelson, 2020; Strazzulli et al., 2017). Metagenomic analysis of “biological dark matter” has significantly extended the entire tree of life and added a substantial number of novel archaeal and bacterial lineages and phyla with no cultivated representatives (Parks et al., 2017; Tahon et al., 2021). The Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org/), based on a comparison of genomes and metagenomes (Parks et al., 2022), currently contains (April 24, 2024; Release 09-RS2020) 175 bacterial and 19 archaeal phylum-level lineages, many of which represent uncultivated organisms. Although representatives of the archaeal domain are ubiquitously distributed, with many novel phyla and lineages having been discovered during the last decade, bacterial representatives are still dominant among the cultivated isolates.
We performed the first metagenomics-based assessment of microbiota in a geothermal spring in Tajikistan. Several high-quality metagenome-assembled genomes (MAGs) were reconstructed, including two archaeal MAGs, one of which represented a novel genus belonging to the hyperthermophilic archaeal family Desulfurococcaceae and phylum Thermoproteota. This family was first described in 1982 as extremely thermophilic, anaerobic, sulfur-respiring, or fermentative archaea that utilized proteinaceous substrates such as yeast extract or casein (Zillig et al., 1982). Desulfurococcaceae currently encompasses 11 validly published genera, with representatives isolated from globally widespread terrestrial hot springs (https://lpsn.dsmz.de/family/desulfurococcaceae), some of which host small obligate parasites belonging to the candidate phylum Nanoarchaeota (Huber et al., 2002; St John et al., 2019; Wurch et al., 2016). Thermophilic archaea have not yet been isolated from the Central Asian region, although a metagenomic survey has identified a few archaea in a hot spring in Kazakhstan (Mashzhan et al., 2021).
2 MATERIALS AND METHODS
2.1 Sampling and sample processing
Water samples were collected from the Tamdykul geothermal field, located 25 km North-Northwest of the Jirgital district center, in the upper part of the Tamdykul valley near Tamdykul river, at an altitude of 2198 masl (GPS location: 39°25.102′ N, 71°13.103′ E; Figures A1 and A2). The conductivity, temperature, and pH of the outlet water were 1019 μS/cm, 88°C, and 7.4, respectively (Dzhuraeva et al., 2021). The sample was transported to the Tajik National University Biotechnology Center in Dushanbe in 5-l sterile plastic bottles and processed immediately via first filtering through a 1.2 mm pore-sized membrane filter to remove debris and particles, followed by filtration through a 0.2 µm pore-sized membrane filter to entrap the cells. The filter membranes were cut into pieces and stored in RNA later (Sigma-Aldrich). Membrane pieces were washed by dipping in 0.85% sodium chloride 3–5 times. DNA was then extracted using a DNeasy PowerSoil Kit (Qiagen), as per the manufacturer's instructions. DNA quality was assessed using a Qubit HS dsDNA kit (Life Technologies) and agarose gel electrophoresis. Sampling was first performed in November 2018 (50 l); however, because the DNA yield was not sufficient for shotgun Illumina sequencing, sampling was repeated in September 2019 (75 l). For filtration and DNA extraction, 120 l water was used. DNA from the two samples were pooled and sequenced. The filtered sample was transported to Bergen, Norway, to analyze major and minor elements and anions using an iCAP™ 7600 ICP-OES Analyzer (Thermo Fisher Scientific) and ion chromatography (IC; Metrohm), respectively, (Table A1) at the ICP-Laboratory at the University of Bergen (https://www.uib.no/en/geo/111639/icp-laboratory). The major cations found were sodium (179.5 ppm), silicon (50.0 ppm), potassium (7.5 ppm), and calcium (3.8 ppm). The major anions found were sulfate (193.2 ppm) and chloride (35.1 ppm). Nitrates, arsenic, cobalt, chromium, copper, europium, iron, lanthanides, vanadium, yttrium, or zinc were not detected.
2.2 Metagenome sequencing and bioinformatics
The metagenome was commercially sequenced at Eurofins Genomics, Constance, Germany (https://eurofinsgenomics.eu/) using HiSeq Illumina technology in the NovaSeq 6000 S2 PE150 XP sequence mode and a NEBNext Ultra II library preparation kit (New England Biolabs). Raw reads were imported to the CLC Genomic Workbench (version 22.0; Qiagen Bioinformatics), paired, and trimmed with Trim Reads followed by taxonomic profiling using the Taxonomic Profiling tool and a custom microbial genome reference database (CLC) downloaded on April 22, 2024. Trimmed reads were assembled and binned using the De Novo Assemble Metagenome and Bin Pangenome by Sequence tools, respectively, as implemented in the Microbial Genomics Module of the CLC Genomics Workbench. Bins were uploaded to KBase (https://www.kbase.us/), quality assessed using CheckM v1.0.18 (Parks et al., 2015), and taxonomically assigned using GTDB-Tk v2.3.2 (Chaumeil et al., 2022). Annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline v. 6.1 (https://www.ncbi.nlm.nih.gov/genome/annotation_prok) and the Rapid Annotation Using Subsystems Technology (RAST) server (https://rast.nmpdr.org) (Aziz et al., 2008). Metabolic features were predicted using BlastKOALA (Kanehisa et al., 2016) and RAST. The secondary structure of the 16S rRNA was predicted using Mfold (Zuker, 2003). A 16S rRNA gene sequence-based phylogenetic tree was constructed using MEGA X (Kumar et al., 2018) and the Maximum Likelihood algorithm. A whole-proteome-based phylogenomic analysis was conducted using FastME 2.1.6.1 (Lefort et al., 2015), employing the Type (Strain) Genome Server (Meier-Kolthoff & Göker, 2019), available at the DSMZ website (https://tygs.dsmz.de). The tree was rooted at the midpoint (Farris, 1972). CRISPR/Cas regions were identified using CISPRCasFinder (Abby et al., 2014; Couvin et al., 2018; Grissa et al., 2007). Genome comparisons using BLAST Ring Image Generator were performed as previously described by Alikhan et al. (2011). Functional annotation to Clusters of Orthologous Groups (COG) was performed using the EggNOG-mapper (Cantalapiedra et al., 2021; Huerta-Cepas et al., 2019).
3 RESULTS AND DISCUSSION
3.1 Metagenomic sequencing, taxonomic profiling, and binning
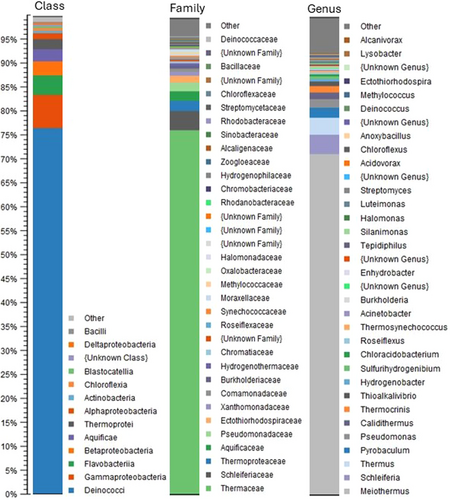
Illumina sequencing of the extracted DNA yielded 9,863,623 reads with a total of 2,959,086,900 sequenced bases. Taxonomic profiling at the Class, Family, and Genus levels using paired reads revealed a high abundance of bacteria belonging to the Deinococci class (>75% abundance), most of which were affiliated with the genus Meiothermus (~70%), a common heterotrophic inhabitant of hot springs that feeds on a large range of carbohydrates, organic acids, and amino acids (Albuquerque et al., 2009); whereas, members of Thermus and Calidithermus constituted 4%–5% of the total population (Figure 1). Pseudomonadota, including Gamma-, Beta-, Alpha-, and Deltaproteobacteria constituted ~10% of the microbial community with a large diversity of genera, with Pseudomonas and chemolithotrophic sulfur oxidizers belonging to genera such as Thioalkalivibrio as dominant representatives. The third largest group was Flavobacteria (~4%), almost exclusively represented by a member of Schleiferia, so far encompassing only one species, Schleiferia thermophila, isolated from a hot spring in the Island of São Miguel in the Azores (Albuquerque et al., 2011). Members of the deep-branching class Aquificia and the archaeal class Thermoprotei constituted approximately 3% and 2%, respectively. Only members of the metabolically versatile Pyrobaculum genus, encompassing both heterotrophic and chemolithoautotrophic hyperthermophilic members with optimal growth temperatures up to 100°C (Huber et al., 1987), were detected among the archaeal group in the taxonomic profiling results. Except for the strong dominance of Meiothermus, the microbial community was similar to that of other circumneutral terrestrial hot springs that have been characterized. Minor numbers of possible phototrophic members were identified, including members of Thermosynechococcus, Chromatiaceae, and Chloroflexia. Class Aquificia was mostly represented by the chemolithotrophic genera Thermocrinis and Hydrogenobacter, common inhabitants of hot springs that oxidize hydrogen, thiosulfate, or elemental sulfur.
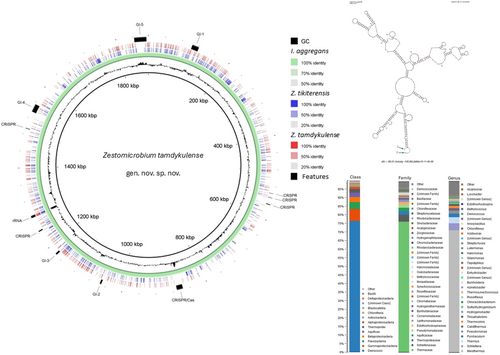
Following trimming and assembly, a total of 19,712,861 unique base pairs of sequence data were obtained and distributed into 1175 contigs with minimum and maximum lengths of 500 and 505,044 base pairs, respectively. Binning resulted in six high-quality MAGs with 88.2%–100% completeness and 0%–2.6% contamination (Table 1). These MAGs covered 82.5% of the total metagenomic sequence data and thus represented most of the microorganisms in the sample. MAGs representing the bacterial taxa Meiothermus cerbereus, Aquificaceae, and Halothiobacillaceae constituted the major components, with a 41.2%, 19.7%, and 18.1% share in the metagenome, respectively, including a minor component of Schleiferia thermophila (1.6%) (Table 1). Two minor MAGs belonging to archaea were obtained: one representing Pyrobaculum, with a 1.2% share of the metagenome, and the other representing Desulfurococcales (MAG002), with only a 0.7% share of the metagenome (Table 1). These archaeal groups, which constitute a minor part of the Tamdykul geothermal spring, are common inhabitants of hot spring waters. The closest placement average nucleotide identity (ANI) value for MAG002 was 80.95%, which was the lowest of the six MAGs, substantially below the 95%–96% value that is recommended as a species boundary (Yoon et al., 2017); therefore, this indicates taxonomic novelty within the Desulfurococcaceae family (Table 1). MAG002 was the only MAG containing a 16S rRNA gene.
| Bin designation | MAG designation | Length (Mbp) | # contigs | N50 | GC content (%) | Completion (%) | Contamination (%) | Closest placement ANI/MSA | 16S rRNA gene | Taxonomy | Share in metagenomea |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bin001 | MAG001 | 2.53 | 19 | 308,548 | 42.9 | 100 | 0 | 99.6/ | 0 | Schleiferia thermophila | 1.6 |
| Bin004 | MAG004 | 2.8 | 109 | 34,641 | 61.6 | 96.6 | 0 | 94.1/ | 0 | Meiothermus cerbereus | 41.2 |
| Bin01 | MAG01 | 1.82 | 59 | 73,502 | 63.4 | 97.7 | 0 | -/94.94 | 0 | Halothiobacillaceae | 18.1 |
| Bin002 | MAG002 | 1.29 | 48 | 52,847 | 41.3 | 98.7 | 2.6 | 80.95/90.34 | 1 | Desulfurococcales | 0.7 |
| Bin007 | MAG007 | 1.4 | 136 | 15,663 | 43.3 | 88.2 | 1.22 | 98.84/ | 0 | Aquificaceae | 19.7 |
| Bin003 | MAG003 | 2.1 | 195 | 16,469 | 55.2 | 99.3 | 0.74 | 99.57/ | 0 | Pyrobaculum sp. | 1.2 |
- a Calculated by mapping raw reads to each individual bin using the Map Reads to Reference tool in CLC.
3.2 Desulfurococcales MAG002
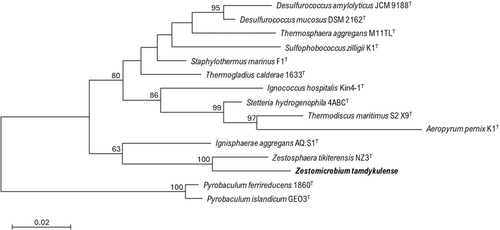
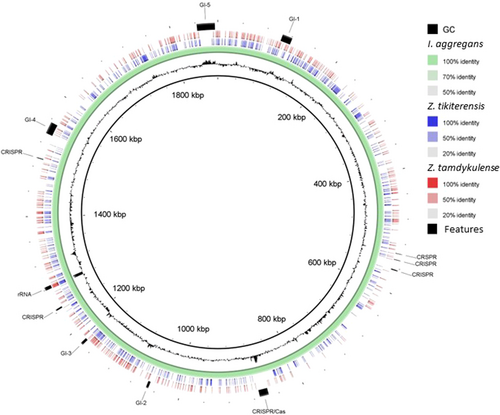
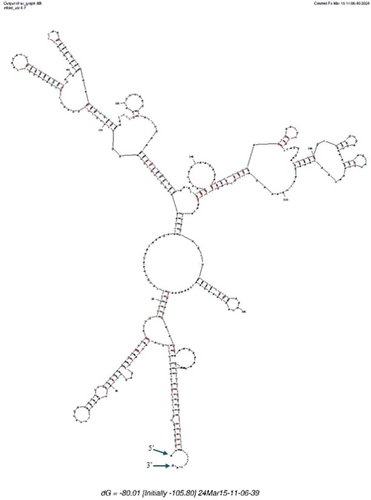
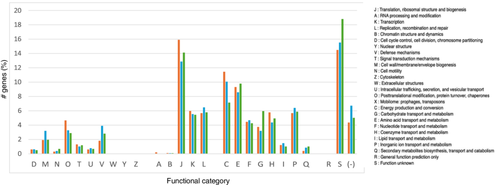
The 16S rRNA gene sequence of MAG002 most closely matched that of Ignisphaera aggregans using BLASTN searches. The gene, containing a small 421-bp intron, was integrated into a large contig. Introns have previously been found in archaeal rRNA genes, including those of Desulfurococcales (Nomura et al., 1998; St John et al., 2019); they sometimes encode homing endonucleases that promote their dissemination through archaeal communities (Aagaard et al., 1995; Jay & Inskeep, 2015). The MAG002 intron did not share any significant sequence similarity with the database entries when analyzed using BLASTN, nor did it encode any homing endonucleases. However, structural prediction revealed potentially extensive secondary structures (Figure A3). After removing the intron from the 16S rRNA gene sequence, the closest BLASTN match was with Zestosphaera tikiterensis NZ3 (MH252993.1), sharing 92.52% sequence identity, indicating that MAG002 represented a novel distinct genus, which was clearly below the recommended 16S rRNA gene sequence threshold value of 94.5% for genera (Yarza et al., 2014). The reconstruction of 16S rRNA- and proteome-based phylogenetic trees, including members of all Desulfurococcaceae genera, placed MAG002 in a lineage between Z. tikiterensis and I. aggregans AQ.S1 (Figures 2 and 3, respectively). Genome sequence comparisons showed the highest digital DNA-DNA hybridization value for I. aggregans (33.7%) and the highest ANI value for Z. tikiterensis (67.7%), supporting taxonomic novelty (Figure 3). A MAG from a Yellowstone National Park hot spring in the USA formed the closest branch, with an ANI value of 77.6% when compared with MAG002 (Figure 3); therefore, it was sufficiently different to constitute a separate genome species or genus. Genome comparisons using the BLAST ring generator with the I. aggregans genome as a reference revealed scattered regions of sequence homology (Figure 4). No CRISPR/Cas regions were found in MAG002, in contrast to the reference strain, AQ.S1, which contained one CRISPR/Cas region and several regions with CRISPR repeats. Genomic islands identified in the reference genome were either absent in MAG002 (e.g., GI–5) or had partial homology (e.g., GI–1 and GI–4). COG analysis revealed a relatively smaller number of genes in the S (function unknown) category in Zestomicrobium tamdykulense as compared with Z. tikiterensis and I. aggregans (Figure A4), which can be explained by a smaller genome of the former organism (1.3 Mbp) in contrast to the 1.8 and 1.9 Mbp genomes of the two latter organisms. This difference in genome size can also explain the larger number of category J (Translation) genes in Z. tamdykulense compared with the two other genomes as the reduction of genome size generally increases the fraction of essential housekeeping functions.

3.3 Metabolic insights from draft genomes
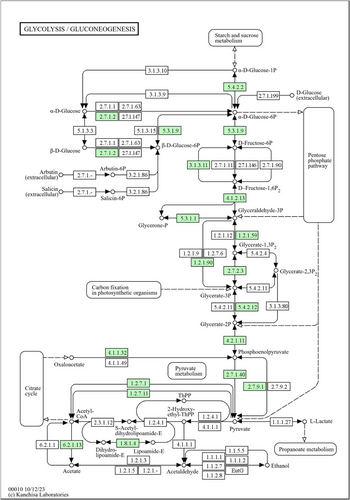
To assess the potential metabolic capabilities of Z. tamdykulense (MAG002), the draft genome sequence was analyzed using BlastKOALA and RAST. The Embden–Meyerhof–Parnas pathway was found to be incomplete, because genes encoding phosphofructokinases, such as 6-phosphofructokinase (EC 2.7.1.11), diphosphate-dependent phosphofructokinase (EC 2.7.1.90), or ADP-dependent phosphofructokinase (EC 2.7.1.146), which convert d-fructose-6-phosphate (F6P) to d-fructose-1,6 P-bisphosphate (FBP), were absent. However, the gene for the bifunctional fructose-1,6-bisphosphate phosphatase/aldolase (EC 3.1.3.11/EC 4.1.2.13) (Aziz et al., 2017; Fushinobu et al., 2011; Say & Fuchs, 2010), which is frequently found in archaea, was identified in MAG002 (Locus tag: QN229_03025). This bisphosphatase dephosphorylates FBP to F6P and also catalyzes the aldolase reaction from FBP to glycerone-phosphate and glyceraldehyde-3-phosphate (Figure A5). The gluconeogenesis pathway is thus complete. However, the bisphosphatase enzyme reaction is irreversible; hence, it renders the glycolysis pathway nonfunctional. Z. tikiterensis has been reported to possess incomplete glycolysis and gluconeogenesis pathways, as it lacks genes for FBP phosphatase and FBP aldolase (St John et al., 2019). However, a BLAST search using the MAG002 protein as a query revealed a partial bisphosphatase gene (expectation value: 2e-69) at the end of a contig in the draft genome sequence of strain NZ3, encoding a truncated 258 amino acid protein (locus tag: PUA32121), which probably represents the bifunctional enzyme.
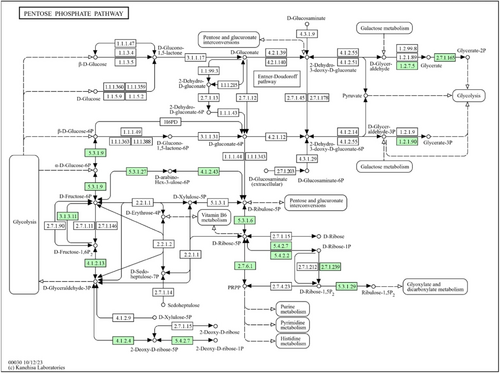
The pentose phosphate pathway, whose major role is to provide 5-carbon sugars for nucleoside biosynthesis, is absent in MAG002 and Z. tikiterensis. However, both genomes encode the ribulose monophosphate pathway enzymes, 6-phospho-3-hexuloisomerase (EC 5.3.1.27; locus tag: QN229_05875) and 3-hexulose-6-phosphate synthase (EC 4.1.2.43; locus tag: QN229_02210), which makes a shunt from F6P via d-arabino-hex-3-ulose-6-phosphate to d-ribulose-5-phosphate when operating in the reverse direction, which can then be converted further to phosphoribosyl pyrophosphate, a central precursor for purine and pyrimidine biosynthesis (Figure A6). Z. tamdykulense assimilates nitrogen from ammonia using glutamate dehydrogenase (EC 1.4.1.2; locus tag: QN229_05895) and the GS/GOGAT system involving sequential action of glutamine synthetase (EC 6.3.1.2; locus tag: QN229_02995) and glutamine oxoglutarate aminotransferase/glutamate synthase (EC 1.4.1.13; locus tag: QN229_01995). The Entner–Doudoroff and the citric acid pathways are absent.
Z. tikiterensis encodes two membrane-bound [NiFe] group 4b hydrogenase clusters, one of which includes a putative catalytic carbon monoxide dehydrogenase subunit (cooS), which suggests energy conservation via carboxydotrophic hydrogenogenesis (St John et al., 2019). The cooS subunit was not detected in MAG002. Another group 4b hydrogenase cluster includes a Na+/H+ antiporter, which is also found in a similar cluster in Z. tamdykulense, suggesting the use of a Na+ gradient for ATP generation using an archaeal-type ATP synthase in both organisms. The Z. tikiterensis genome also encodes a NiFe 3 A hydrogenase cluster group containing a coenzyme F420-reducing hydrogenase subunit homolog (St John et al., 2019), normally involved in methanogenesis. However, as the F420 biosynthetic pathway is not found in these organisms, this enzyme probably uses another cofactor. A homolog sharing 28% amino acid identity was also detected in Z. tamdykulense, but this belonged to a cluster of four genes annotated as sulfhydrogenases (Locus tags: QN229_02275, QN229_02280, QN229_02284, QN229_02290). Sulfhydrogenases are bifunctional 4-subunit enzyme complexes that evolve hydrogen and reduce elemental sulfur to hydrogen sulfide (EC 1.12.1.5 and EC 1.12.98.4). This enzyme is important for fermentation in the hyperthermophilic archaeon Pyrococcus furiosus (Adams et al., 2001).
Z. tamdykulense is an amino acid auxotroph as none of the amino acid biosynthesis pathways seem to be complete. This is also the case for purines and pyrimidines except for uracil biosynthesis. Two ABC nucleoside uptake gene clusters and two clusters of oligopeptides/dipeptides ABC transporter systems were identified in the metagenome. A total of 22 genes encoding ATP-binding proteins for the transport of oligopeptides, dipeptides, and amino acids were found, indicating a high potential for importing a large variety of organic nutrients, adapting to growth in nutrient-rich environments, and supporting an auxotrophic lifestyle.
4 CONCLUSIONS
Metagenomic analysis of the Tamdykul geothermal spring revealed a high abundance of bacteria from the families Thermaceae, Aquificaceae, and Halothiobacillaceae, along with several high-quality MAGs, including one from a novel genus of the archaeal family Desulfurococcaceae. Further analysis of this MAG revealed a 16S rRNA gene sequence containing a 421 bp noncoding intron. The MAG encodes a complete gluconeogenesis pathway involving a bifunctional fructose-1,6-bisphosphate phosphatase/aldolase; however, the glycolysis pathway is incomplete. It contains ribulose monophosphate pathway enzymes, which may be used for pentose synthesis. Additionally, it encodes several hydrogen-evolving hydrogenases with possible roles in fermentation as hydrogen sinks. We propose the name Z. tamdykulense gen. nov., sp. nov. for this organism, which is the first reported thermophile genome from Tajikistan. The thermophile genomes presented in this work represent a rich source of novel thermostable enzymes with potential use in agro-industrial processes and molecular biotechnology.
AUTHOR CONTRIBUTIONS
Munavvara Dzhuraeva: Conceptualization (equal); data curation (lead); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); visualization (equal); writing—original draft (equal); writing—review and editing (equal). Khursheda Bobodzhanova: Conceptualization (equal); data curation (supporting); formal analysis (supporting); funding acquisition (equal); investigation (supporting); methodology (supporting); project administration (equal); supervision (equal); validation (supporting) writing—original draft (supporting); writing—review and editing (supporting). Nils-Kåre Birkeland: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (equal); investigation (equal); methodology (supporting); project administration (lead); supervision (equal); validation (equal); writing—original draft (equal); writing—review and editing (lead).
ACKNOWLEDGMENTS
This research was funded by the Eurasia Program of the Norwegian Directorate for Higher Education and Skills (HK-dir) under Project CPEA-LT-2017/10061. The authors are grateful to the ICP-Laboratory at the University of Bergen for their excellent service in performing the chemical analyses for this study.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
None required.
APPENDIX

| Elements | (ppm) | Anions | (ppm) |
|---|---|---|---|
| Aluminum | 0.035 | Bromide | 0.92 |
| Boron | 1.324 | Chloride | 35.10 |
| Barium | 0.004 | Sulfate | 193.17 |
| Calcium | 3.797 | ||
| Lead | 0.018 | ||
| Lithium | 0.595 | ||
| Magnesium | 0.088 | ||
| Manganese | 0.008 | ||
| Nickel | 0.015 | ||
| Sodium | 197.480 | ||
| Strontium | 0.119 | ||
| Sulfur | 83.039 | ||
| Silicon | 50.028 | ||
| Titanium | 0.001 | ||
| Potassium | 7.503 |
Open Research
DATA AVAILABILITY STATEMENT
The nucleotide sequence of MAG002 reported and discussed in this paper is deposited at DDBJ/ENA/GenBank under accession JAVVFV000000000: https://www.ncbi.nlm.nih.gov/nuccore/JAVVFV000000000. The version described in this paper is JAVVFV010000000. Zestomicrobium tamdykulense SeqCode registry accession: https://registry.seqco.de/registers/r:9-vfcl8f.