Responses of Anabaena sp. PCC7120 to lindane: Physiological effects and differential expression of potential lin genes
Graphical Abstract
In this article, the tolerance of Anabaena sp. to lindane was studied as well as the expression of its potential lin genes. Our goal was to obtain information to create strategies for lindane bioremediation and also to find the induction of genes in the presence of lindane that can be used in the development of whole-cell biosensors.
Abstract
Lindane (γ-HCH) is an organochlorine pesticide that causes huge environmental concerns worldwide due to its recalcitrance and toxicity. The use of the cyanobacterium Anabaena sp. PCC 7120 in aquatic lindane bioremediation has been suggested but information relative to this process is scarce. In the present work, data relative to the growth, pigment composition, photosynthetic/respiration rate, and oxidative stress response of Anabaena sp. PCC 7120 in the presence of lindane at its solubility limit in water are shown. In addition, lindane degradation experiments revealed almost a total disappearance of lindane in the supernatants of Anabaena sp. PCC 7120 culture after 6 days of incubation. The diminishing in lindane concentration was in concordance with an increase in the levels of trichlorobenzene inside the cells. Furthermore, to identify potential orthologs of the linA, linB, linC, linD, linE, and linR genes from Sphingomonas paucimobilis B90A in Anabaena sp. PCC 7120, a whole genome screening was performed allowing the identification of five putative lin orthologs (all1353 and all0193 putative orthologs of linB, all3836 putative orthologs of linC, and all0352 and alr0353 putative orthologs of linE and linR, respectively) which could be involved in the lindane degradation pathway. Differential expression analysis of these genes in the presence of lindane revealed strong upregulation of one of the potential lin genes of Anabaena sp. PCC 7120.
1 INTRODUCTION
Hexachlorocyclohexane (HCH) was widely used as a pesticide between 1930 and 1990, being first applied as a technical mixture that contained α-HCH, β-HCH, γ-HCH, δ-HCH, and ε-HCH. HCH is usually synthesized by benzene photochlorination, a process in which the aforementioned isomers were formed. Afterward, solely lindane (γ-HCH isomer) was commercialized because, among all the isomers, only γ-HCH had strong insecticidal properties (Li, 1999). Since the industrial procedure rends only about 10%–15% of γ-HCH, it has been estimated that, in those years, between four and six million tons of the other HCH isomers were dumped worldwide generating an important environmental and health concern (Lal et al., 2010). In 2009, HCH isomers were defined as persistent organic pollutants in the Stockholm Convention (2009) and since 2015 lindane was included in the list of carcinogenic agents for humans by the World Health Organization (2018). Although HCH is durable and recalcitrant, some microorganisms are capable of degrading one or more HCH isomers (Lal et al., 2010). The best characterized in terms of their lindane biodegradation abilities are heterotrophic bacteria belonging to the Sphingomonadaceae family (Verma et al., 2014). The diversity, organization, and distribution of lindane catabolic pathways are reasonably well-established in these bacteria (Verma et al., 2014). Aerobic lindane degradation pathways are conformed by lin genes and are subdivided into upper and lower pathways. In particular, the lin pathways are well-characterized in Sphingobium japonicum UT26S (Nagata et al., 1999, 2007) and Sphingobium indicum B90A (Lal et al., 2010). The upper pathway comprises linA (HCH dechlorinase), linB (haloalkane dehalogenase), and linC (dehydrogenase) whereas the lower pathway is formed by linD (reductive dechlorinase), linE (ring cleavage oxygenase), linF (maleylacetate reductase), linG,H (acyl-CoA transferase), linJ (thiolase) and linI and linR (transcriptional regulators) (Nagata et al., 2007; Verma et al., 2014). The dynamics of lin genes expression in Sphingomonas paucimobilis B90A (also known as S. indicum B90A) revealed that linA, linB, and linC were expressed constitutively whereas, on the contrary, the expression of linD and linE was induced in presence of 7 mg/L of lindane and 2 mg/L of α-HCH (Suar et al., 2004).
Among photoautotrophic organisms, Anabaena sp. PCC 7120, Nostoc ellipsosporum, and Microcystis aeruginosa PCC 7806 have been described as potential lindane degraders (Ceballos-Laita et al., 2015; Kuritz & Wolk, 1995; Kuritz et al., 1997; Sarasa-Buisán et al., 2022). Anabaena sp. PCC 7120, a nitrogen-fixing cyanobacterium, was reported to be able to metabolize lindane (γ-HCH) yielding pentachlorocyclohexene and 1,2,4-trichlorobenzene in a process dependent on nir operon, which encodes some enzymes for nitrate utilization (Kuritz & Wolk, 1995; Kuritz et al., 1997). However, the genes and subsequent catabolic pathways involved in such a process remain unknown. In addition, the effect of the presence of lindane on the photosynthetic apparatus of Anabaena sp. PCC 7120 was analyzed, concluding that photosynthetic activity was slightly increased in the presence of lindane and that no changes were observed in the synthesis and activity of ferredoxin-NADP+ reductase (FNR) (Bueno et al., 2004).
On the other hand, it was previously reported that the levels of lindane in supernatants of M. aeruginosa PCC 7806 cultures treated with 7 mg/L lindane decreased significantly after 15 days of treatment (Ceballos-Laita et al., 2015). Nevertheless, only an ortholog of the linC gene was found in the genome of M. aeruginosa NIES-843, which interestingly was induced in the presence of lindane (Sarasa-Buisán et al., 2022).
The lin pathway probably constitutes a unique biochemical system that allows HCH detoxification. Until the moment the genes that constitute this pathway and the substrate specificity, isomer coverage, and kinetics of enzymes are relatively well-known in Sphingomonads. However, information concerning this pathway in other organisms is scarce (Lal et al., 2010). The knowledge of the presence of lin genes as well as the tolerance of Anabaena to lindane is important to evaluate the potential use of this model cyanobacterium in bioremediation strategies or in other processes such as the development of whole-cell biosensors to detect HCH isomers.
In this work, Anabaena sp. PCC 7120 physiological responses to the presence of lindane, namely growth rate, pigment composition, photosynthetic/respiration rates, and oxidative stress response were evaluated. Moreover, a search of lin genes in the genome of this cyanobacterium was performed and their transcriptional responses in the presence of lindane were studied. Finally, Anabaena sp. PCC 7120 degradative capacity was also studied by testing the disappearance of lindane in the supernatants of cells treated with this pesticide as well as the appearance of lindane metabolic intermediaries inside Anabaena sp. PCC 7120 cells.
2 MATERIALS AND METHODS
2.1 Strains and culture conditions
Anabaena sp. PCC 7120 was grown photoautotrophically in BG-11 medium (Stanier et al., 1979) containing 30 μM of FeSO4 instead of 30 μM of ammonium ferric citrate to avoid inhibition of lindane metabolization (Kuritz & Wolk, 1995). These cultures were maintained in 250 mL Erlenmeyer flasks holding 100 ml of Anabaena sp. PCC 7120 cultures each at 28°C in a New Brunswick™ Innova® 43 Shaker under continuous illumination with white light at 30 μmol photons m−2 s−1 and gentle shaking at 100 rpm. To study the effects of lindane (γ-HCH) on Anabaena sp. PCC 7120, 10 μL of a solution of 70 mg/mL of γ-HCH in DMSO were added to 250 mL Erlenmeyer flasks containing 100 mL of culture, obtaining a final concentration of 7 mg/L. The effect of DMSO on Anabaena sp. PCC 7120 was also analyzed by adding 10 μL of DMSO to 250 mL Erlenmeyer flasks containing 100 mL of culture.
2.2 Growth measurement
The effect of 7 mg/L of γ-HCH on the photoautotrophic growth of three independent cultures of Anabaena sp. PCC 7120 with an initial OD750nm of 0.3 was measured spectrophotometrically using a Cary 100 Bio UV-Visible spectrophotometer (Varian). The optical density was recorded at 750 nm for 22 days every 48 h. At the same time, the packed cell volume (PCV) was measured using 5 mL graduated tubes with a capacity of 60 μL of packed volume. Five milliliters of cultures were centrifuged in the aforementioned tubes for 5 min at 18°C and 2000xg using an Allegra X 30 R centrifuge. PCV measurements were expressed as microliters of PCV per milliliter of culture.
2.3 Pigment content determinations
Pigment content of Anabaena sp. PCC 7120 in the presence of 7 mg/L of γ-HCH was determined at exponential (OD750nm = 1.0) and stationary (OD750nm = 2.0) phases of growth in three independent cultures of each condition with an initial OD750nm of 0.3. Chl a, phycobiliprotein, and carotenoid levels were quantified as described by Mackinney (1941), Glazer (1976), and Davies (1976), respectively.
2.4 Net photosynthesis and dark respiration rate measurements
Net photosynthesis and dark respiration rates of Anabaena sp. PCC 7120 were measured at room temperature with a Clark-type oxygen electrode model Chlorolab 2 (Hansatech). Net photosynthesis (defined as true photosynthesis minus photorespiration and dark respiration (Wohlfahrt & Gu, 2015), was determined by measuring the O2 increase during 5 min illuminating cell suspensions with white light at 10, 50, 100, and 400 µmol photons·m−2·s−1. Dark respiration was determined by measuring the consumption of oxygen during 3 min in darkness. Both rates were measured at exponential (OD750nm = 1.0) and stationary (OD750nm = 2.0) phases of growth and were expressed as pmol O2 × s−1/μL of PCV. All measurements were performed in triplicate.
2.5 Catalase and superoxide dismutase (SOD) activity determinations
Catalase and SOD activities were measured after 48 h of exposure to 7 mg/L of lindane in three independent cultures with an initial OD750nm of 0.3. 50 mL of cell culture were centrifuged at 3000 g and 4°C for 10 min and cells were resuspended in 1 mL of phosphate buffer (KH2PO4/K2HPO4) 50 mM pH 7. Then cells were broken by sonication, the resulting solution was centrifuged at 13,000 g and 4°C for 10 min and the total protein concentration was determined in the supernatant by using the BCA™ Protein Assay kit (Thermo Fisher Scientific).
Catalase activity was determined as described by Beers and Sizer (1952). Six hundred micrograms of protein extract was mixed with H2O2 to a final concentration of 20 mM and the dissociation of H2O2 was followed at 240 nm with a Cary 100 Bio UV-Visible spectrophotometer (Varian) for 5 min. Catalase activity was expressed in units per milligram of total protein, defining a unit as the amount of enzyme that dissociates 1 μg of H2O2 per minute.
SOD activity was measured as described by Winterbourn et al. (1975) with some modifications implemented by Sein-Echaluce et al. (2015). Six hundred micrograms of protein extract was mixed with 6.4 mM ethylenediaminetetraacetic acid (EDTA), 41 μM nitro-blue tetrazolium (NBT), 2.3 μM riboflavin and 23.5 μM N,N,N′,N′-Tetramethylethylenediamine (TEMED) and SOD activity was determined by measuring the inhibition of NBT reduction by SOD. Thus, absorbance at 560 nm was determined before and after illuminating the mixtures for 10 min with UV light, using a control containing phosphate buffer instead of the protein extract. SOD activity was expressed in units per milligram of total protein, defining a unit as the amount of enzyme that inhibits the maximum reduction to half.
2.6 Microscopy experiments
To analyze the effects of lindane on the morphology of Anabaena sp. PCC 7120 filaments, bright-field, and fluorescence microscopic examinations were carried out with a Nikon Eclipse 50i Epi-fluorescence microscope. Photographs were taken with a Nikon DXM1200F camera coupled to the microscope both at exponential (OD750nm = 1.0) and stationary (OD750nm = 2.0) phases of growth. Fluorescence was detected using a 560/40 nm excitation filter, a 595 nm dichroic beam splitter, and a 630/60 nm emission filter.
2.7 Identification of putative lin genes in the genome of Anabaena sp. PCC 7120
To identify putative lin genes in the genome of Anabaena sp. PCC 7120 the sequences of the proteins encoded by genes linA, linB, linC, linD, linE, and linR from S. paucimobilis B90A were retrieved from Uniprot (https://www.uniprot.org/). Using these sequences as queries and an expectancy-value threshold of 0.005, a protein BLAST (Basic Local Alignment Search Tool) using the CyanoBase Similarity Search (http://genome.microbedb.jp/blast/blast_search/cyanobase/genes) was performed on Anabaena sp. PCC 7120 genome. In all cases, the sequence with the lowest expected value was selected and a pairwise global alignment using ClustalW (https://www.genome.jp/tools-bin/clustalw) was performed using default settings.
2.8 Degradation measurements
The degradation of lindane was studied in three independent cultures with an initial OD750nm of 0.6 containing 7 mg/L of lindane. Erlenmeyer flasks containing 7 mg/L of lindane in BG-11 medium without cells served as controls for evaporation and photodegradation. After 1, 3, and 6 days of treatment, 10 mL of each sample was taken and centrifuged for 5 min at 3000 g in the case of Anabaena sp. PCC 7120 cultures. Lindane analysis of these samples was carried out in the HCH Analysis laboratories at the facilities of the new security cell in Bailín II (Sabiñánigo, Huesca). The isomers of HCH in an aqueous matrix were extracted using a liquid-liquid extraction with hexane and the quantification of the remaining amount of each isomer was carried out using gas chromatography with a mass detector (Agilent Series 7890 A). The identification of isomers was carried out using commercial individual standards.
The formation of degradation intermediaries was studied in three independent cultures with an initial OD750nm of 0.6 containing 7 mg/L of lindane. Ten milliliters of cultures were taken after 0 (control), 24, and 48 h of exposure to lindane and then centrifuged for 5 min at 3000 g. A pellet of cells was extracted by adding 2 mL of hexane at 4°C and shaking in a vortex for 5 min. Finally, the resulting suspension was centrifuged at 12,000 g and 4°C for 10 min and the supernatant was used to determine the presence of lindane degradation intermediaries. One microliter of diluted samples were analyzed in a gas chromatography coupled to a mass detector using a 436GC Chromatograph equipped with a Rxi-5Sil MS column (30 m × 0.25 mm × 0.25 μm). Gas chromatography/mass spectrometry (GC-MS) analyses were performed at the Interdepartmental Research services of the Autonomous University of Madrid, Spain.
2.9 RNA extraction and real-time RT-PCR
The effects of lindane on gene expression were analyzed in three independent cultures of Anabaena sp. PCC 7120 with an initial OD750nm of 0.3. RNA was extracted from 25 mL of each culture after 12 and 24 h of exposure to 7 mg/L of lindane following a method described in Sarasa-Buisan et al. (2022). The absence of DNA in the RNA samples was checked by real-time PCR, using oligonucleotides for the housekeeping gene rnpB (Vioque, 1992). RNA was quantified spectrophotometrically using a SPECORD® PLUS Analytik Jena spectrophotometer.
Two micrograms of total RNA was reverse-transcribed using SuperScript retrotranscriptase (Invitrogen) following the manufacturer's conditions. Real-time PCR was performed using the ViiA™ 7 real-time PCR System (Applied Biosystems). The specific primers for each gene that were analyzed are included in Appendix: Table A1. Each reaction was set up by mixing 12.5 µL of SYBR Green PCR Master Mix with 0.4 μL of 25 µM primer mixture and 10 ng of cDNA template in a final volume of 30 µL. The extension of PCR products was performed at 60°C. The relative mRNA levels of the target genes were normalized to the housekeeping gene rnpB (Vioque, 1992). Relative quantification was performed according to the comparative Ct method (ΔΔCt Method) (Livak & Schmittgen, 2001). The minimum fold-change threshold was set up to ±1.5-fold.
2.10 Statistical tools
To determine whether the changes observed in the measurements of different parameters were significant, t-test statistical analyses were performed by using the GraphPad Prism 7 program.
3 RESULTS
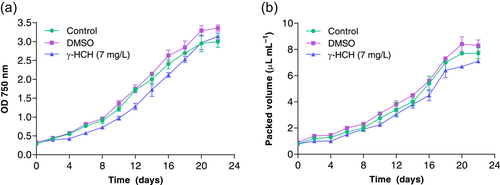
3.1 Anabaena sp. PCC 7120 cells exhibited a high level of tolerance to lindane
Anabaena cells were exposed to 7 mg/L of lindane for 22 days in batch cultures. The concentration added to Anabaena cultures corresponded to the solubility limit of lindane in water. The growth rate was measured by using the optical density at 750 nm as well as the PCV (Figure 1a,b). Since DMSO was used to solve lindane, a control culture in which DMSO alone was added to the cells was monitored. Growth curves representing both parameters, OD at 750 nm and PCV showed similar growth of Anabaena in the presence and absence of lindane (Figure 1a,b). Indeed, the doubling time was 5.28 ± 0.051 d−1 for Anabaena sp. PCC 7120 in the absence of lindane and 5.02 ± 0.47 d−1 in the presence of lindane. t-test statistical analysis of these data revealed that the change observed in the doubling time in the presence of lindane is not significant (p-value = 0.3948).
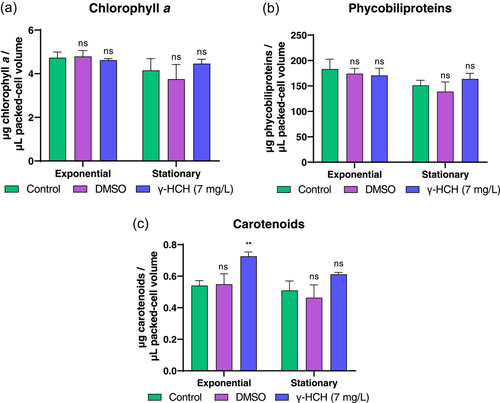
To better understand the physiological responses of Anabaena sp. PCC 7120 to lindane, pigment composition (chlorophyll a, phycobiliprotein, and carotenoid contents) was determined in both exponential and stationary growth phases. The raw values of pigment content were normalized by using the PCV. As can be seen in Figure 2a,b no changes were observed in the level of chlorophyll a and phycobiliproteins present in cells treated with lindane related to those in nontreated cells. However, the level of carotenoids showed a moderated increase in cells treated with lindane in exponential phase, although afterward in stationary phase they remained similar to the untreated cultures (Figure 2c). Taken together, these data indicate that lindane at 7 mg/L concentration barely affects the growth and pigment composition of Anabaena sp. PCC 7120.
3.2 The presence of lindane altered the photosynthetic electron transport and respiration of Anabaena sp. PCC 7120 cells
The photosynthetic rate was measured by using different light thresholds (10, 50, 100, 400 μmol photons m−2s−1) in Anabaena cultures grown in the presence and absence of lindane. Results revealed that the presence of lindane increased the photosynthetic rate in Anabaena cells in both exponential and stationary growth phases under the four conditions tested (Figure 3a–d). Nevertheless, the respiration rate decreased in the lindane-treated cells in exponential growth phase whereas no changes were observed in stationary growth phase (Figure 3e).
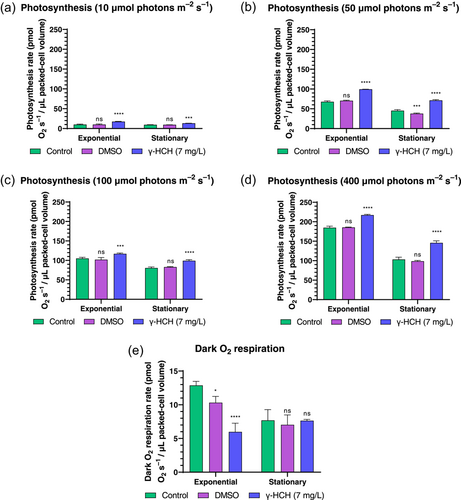
3.3 Lindane triggered oxidative stress in Anabaena cells
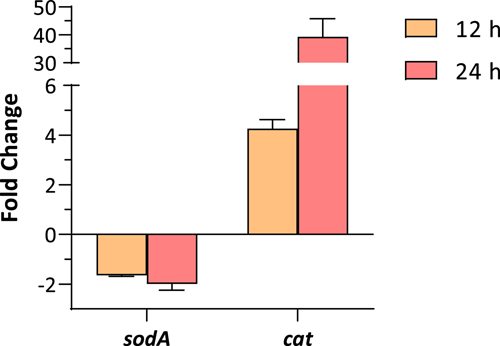
Lindane has been described as a cause of oxidative stress in many organisms, including cyanobacteria (Ceballos-Laita et al., 2015; Deng et al., 2022; Yu et al., 2020). To study whether lindane can induce oxidative stress response, the expression of two key enzymes involved in the detoxification of reactive oxygen species (ROS) was analyzed in Anabaena sp. PCC 7120 culture after 12 and 24 h of treatment with 7 mg/L of lindane. The selected enzymes were superoxide dismutase A (sodA) and catalase (cat). Results shown in Figure 4 indicated that the expression of sodA was slightly downregulated but the expression of cat was strongly upregulated in these conditions (four-fold after 12 h of lindane treatment and 40-fold after 24 h of lindane treatment). In addition, superoxide dismutase and catalase activities were also determined in cultures after their exposure to 7 mg/L of lindane. Results are shown in Table 1. Catalytic activities were in agreement with transcriptional data since a significant increase in catalase activity was observed in cells exposed to lindane whereas the SOD activity was not altered in these conditions (Table 1). In summary, these results strongly suggest the induction of the oxidative stress response in Anabaena sp. PCC 7120 cells as a consequence of the presence of lindane.
| Enzymatic activities (U/mg total protein) | Anabaena sp. PCC 7120 | Anabaena sp. PCC 7120 + 7 mg/L γ-HCH |
|---|---|---|
| Superoxide dismutase activity | 2.93 ± 0.13 | 2.74 ± 0.05ns |
| Catalase activity | 11.01 ± 1.03 | 24.4 ± 2.1* |
- Note: Enzymatic activities were subjected to a t-test with respect to the control to determine if values were significant: ns, not significant.
- * p < 0.05.
3.4 Anabaena sp. PCC 7120 cells were able to metabolize lindane
Lindane degradation experiments were performed with Anabaena PCC 7120 by using a BG11-modified media lacking ammonia to avoid inhibition of lindane metabolization (Kuritz & Wolk, 1995). Cultures were incubated with 7 mg/L of lindane in triplicated and aliquots of the supernatants were taken at different times to determine the decrease in lindane content. Data shown in Figure 5a indicated that Anabaena was able to metabolize lindane almost totally after 6 days of the experiment. The degradation seemed to start after 1 day of exposure to lindane and its concentration decreased to 40.43% after 3 days of incubation and 99.75% after 6 days of incubation (Figure 5a).
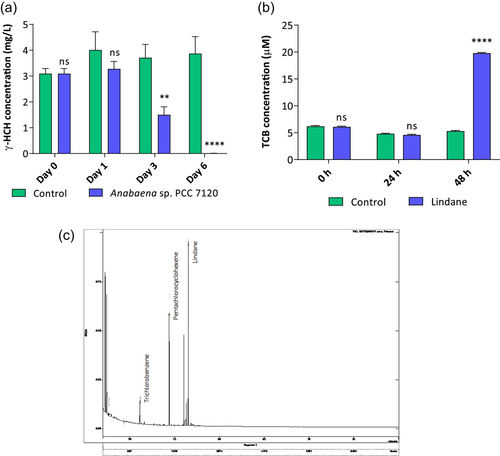
The presence of some metabolic intermediaries of the lindane degradation pathway of S. paucimobilis was analyzed by GC-MS inside of the Anabaena sp. PCC 7120 cells were treated with lindane after 24 and 48 h of treatment. The intracellular intermediates analyzed were pentachlorocyclohexene and trichlorobenzene (resulting products of LinA enzyme), 2-5-dichlorophenol (resulting product of LinB enzyme), 2,5-dichlorohidroquinone (resulting product of LinC enzyme) and chlorohydroquinone (resulting product of LinD enzyme) (Nagata et al., 2007). Trichlorobenzene and pentachlorocyclohexene were detected in the cells after 48 h of lindane treatment (Figure 5b,c). However, only TCB was quantified because pentachlorocyclohexene probably is the product of a first step of dechlorination that in turn is transformed into trichlorobenzene in a second step of dechlorination as happens in the first step of lindane degradation performed by linA enzyme in S. paucimobilis. The intermediaries 2-5-dichlorophenol, 2,5-dichlorohidroquinone, and chlorohydroquinone were not found inside Anabaena sp. PCC 7120 cells.
3.5 Anabaena genome contains some putative lin genes
Orthologs of lin genes from S. paucimobilis were searched into the genome of Anabaena PCC 7120 by performing protein BLAST analyses (Table 2). In the case of linB, (haloalkane dehalogenase) two possible orthologs were found. The gene of Anabaena sp. PCC 7120 with the highest similarity was all1353 (41% similarity), which was then called linB1 (Appendix: Figure A1a). However, another ortholog (all0193) was also found. Its similarity, albeit lower, was also significant (33%) but was interestingly annotated as a haloalkane dehalogenase, and was then called linB2 (Appendix: Figure A1b). These in silico analyses also showed a possible ortholog for the gene linC (2,5-dichloro-2,5-ciclohexadiene-1,4-diol dehydrogenase), the gene all3836, exhibiting a 50% of similarity (Appendix: Figure A1c). Interestingly, a S. japonicum UT26 linC ortholog was previously described in M. aeruginosa NIES-834 and a phylogenetic analysis of the presence of linC in cyanobacteria also found all3836 as the putative linC gene in Anabaena sp. PCC 7120 (Sarasa-Buisán et al., 2022). Protein sequence alignments of LinE (hydroquinone 1,2-dioxygenase) and LinR (transcriptional regulator) of S. paucimobilis displayed some degree of homology with the genes products of all0352 and alr0353 from Anabaena PCC 7120, respectively (26.2% in the case of LinE and 46.7% in the case of LinR) (Appendix: Figure A1d,e). However, the similarity between LinE from S. paucimobilis and All0352 from Anabaena sp. PCC 7120 is slightly low (26.2%), it is divergently located with respect to alr0353, which is in agreement with linR and linE arrangement in the Sphingomonas genome, suggesting that both genes could be orthologs of linR and linE. Finally, these in silico approaches did not allow the identification of orthologs for linA (HCH dehydrochlorinase) and linD (2,5-dichlorohidroquinone dechlorinase) in Anabaena sp. PCC 7120 genome.
| Sphingomonas paucimobilis B90A | Anabaena sp. PCC 7120 | Pairwise alignment | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Annotation | Protein length | Gene | Annotation | Protein length | Identity | Similarity | Gaps |
| linA | Gamma-hexachlorocyclohexane dehydrochlorinase | 156 aa | _ | _ | _ | _ | _ | |
| linB | Haloalkane dehalogenase | 296 aa | all1353 | Putative hydrolase | 282 aa | 23.5% | 40.6% | 6.04% |
| all0193 | Haloalkane dehalogenase | 292 aa | 17.5% | 33.3% | 5.94% | |||
| linC | 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase | 250 aa | all3836 | Glucose 1-dehydrogenase | 269 aa | 30.6% | 49.8% | 8.5% |
| linD | 2,5-dichlorohydroquinone reductive dechlorinase | 346 aa | _ | _ | _ | _ | _ | |
| linE | Chlorohydroquinone/hydroquinone 1,2-dioxygenase | 321 aa | all0352 | NADPH-dependent carbonyl reductase | 248 aa | 11.7% | 26.2% | 24.9% |
| linR | HTH-type transcriptional regulator LinR | 303 aa | alr0353 | Transcriptional regulator | 184 aa | 25.5% | 46.7% | 1.3% |
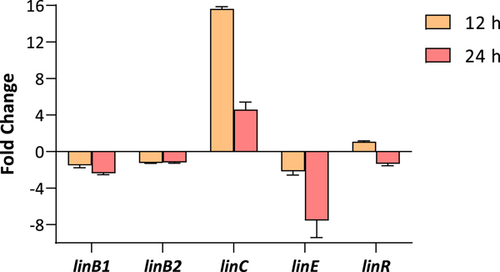
3.6 Dynamics of expression of putative lin genes in Anabaena PCC 7120
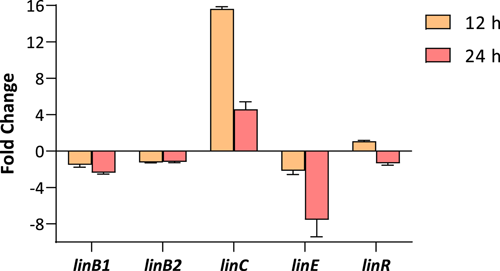
Given the presence of some putative lin genes in the genome of Anabaena sp. PCC 7120, the expression of linB1, linB2, linC, linE, and linR was analyzed by using Real-Time RT-PCR. Anabaena sp. PCC 7120 cells were exposed to 7 mg/L of lindane and gene expression was analyzed after 12 and 24 h of treatment. As can be seen in Figure 6, a clear induction of linC (16-fold) was observed after 12 h of treatment. Nevertheless, after 24 h of treatment, the induction was softened to four-fold. On the contrary, the levels of expression of linE decreased after 24 h of exposure. The transcription of the rest of the genes (linB1, linB2, and linR) remained unaltered.
4 DISCUSSION
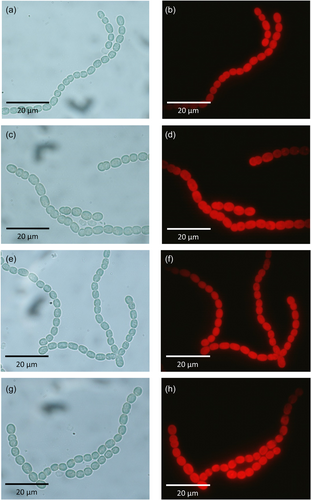
Anabaena sp. PCC 7120 is a model cyanobacterium that exhibits wide biotechnological applications (Abed et al., 2009). The use of Anabaena sp. PCC 7120 in bioremediation processes such as bioaugmentation of lindane-contaminated waters is conditioned by several factors including its resistance to the presence of this organochlorine pesticide. In the present work, we show data relative to the growth, pigment composition, and photosynthetic/respiration rate of Anabaena sp. PCC 7120 in the presence of lindane at its solubility limit in water. Our results indicate that 7 mg/L of lindane barely affects the growth and pigment composition of Anabaena sp. PCC 7120, suggesting a suitable physiology state of the cells under these conditions. This fact is in agreement with no morphological differences observed in Anabaena sp. PCC 7120 cultures treated with lindane both in exponential and stationary growth phases (Appendix: Figure A2). However, the enrichment of antenna complexes in carotenoids is a well-established process that allows the dissipation of free radicals generated by oxidative stress in cyanobacteria (Xiao et al., 2011). Thus, although cells seem not to be affected by lindane, the increase in carotenoid content joined to the strong upregulation of catalase expression suggest that lindane is triggering the oxidative stress response in Anabaena cells.
Surprisingly, we observed an increase in the photosynthetic rate in Anabaena cells treated with lindane. Although these data had been previously reported (Bueno et al., 2004), these results seem to be quite intriguing. One possible explanation could be the fact that as the measurement of photosynthesis is performed determining the release of oxygen, other processes different from photosynthesis could be triggering oxygen production in the presence of lindane, for example, the oxidative stress response via catalase. This hypothesis would be congruent with our observation of upregulation in the expression of catalase and the higher activity of catalase in the cells in the presence of lindane. On the other hand, the data on the decrease in the respiration rate in cells treated with lindane is highly interesting since it might suggest that Anabaena cells are not utilizing lindane as a carbon and energy source as happens in Sphingomonas cells (Nagata et al., 1999), at least in the conditions tested in the present work. It is important to note that Anabaena is a photoautotrophic organism and, under light conditions, this organism uses preferably photosynthetic-derived carbon sources for growth. However, the degradation of lindane by Anabaena grown under dark conditions has never been studied. On the other hand, some bacteria uptake accidentally compounds that they would not metabolize, but if they have enzymes with a low degree of specificity for their main substrates, these compounds can be transformed by these enzymes in other products, and this could be the case of Anabaena and lindane. Finally, the reason why the respiration rate decreased in Anabaena cells in the presence of lindane is unknown but maybe the alterations in the expression of enzymes that carry out the oxidative stress response in the darkness can be modifying the consumption of oxygen as happened in the production of oxygen by photosynthesis in the presence of lindane.
Regarding lindane metabolization, in this work, we observed that lindane content decreased in supernatants of Anabaena sp. PCC 7120 cultures. Simultaneously an increase in the presence of trichlorobenzene is observed inside Anabaena sp. PCC 7120 cells. These data are in agreement with previous results reported by Kuritz and Wolk (1995), but the difference relies on the amount of lindane added to the cultures. In this work, 7 mg/L of lindane was added to the cultures, whereas in the experiments performed by Kuritz and Wolk, assays were carried out by using 0.5 μg/L of lindane. In this work, the authors found an increase in γ-pentachlorocyclohexene and 1,2,4-trichlorobenzene inside the cells treated with lindane. Since both metabolites disappeared over time, our interest was to find other metabolic intermediaries inside Anabaena cells that were similar to those of the Sphingomonas lindane degradation pathway. However, this task was unsuccessful because 2-5-dichlorophenol (resulting product of LinB enzyme), 2,5-dichlorohidroquinone (resulting product of LinC enzyme), and chlorohydroquinone (resulting product of LinD enzyme) were not detected inside Anabaena cells. Intriguingly, the protein sequence similarity searches of lin genes indicated that Anabaena contains two putative orthologs of linB (linB1 and linB2) and linC genes so that the enzymatic reactions catalyzed by these enzymes, as well as the degradation pathway after TCB remains unknown. According to our search of putative lin genes, an ortholog of linA (HCH dechlorinase) was not found in the genome of Anabaena sp. PCC 7120. In Sphingomonas two initial dechlorination reactions carried out by linA produce 1,3,4,6-tetrachloro-1,4-cyclohexadiene which presumptively is spontaneously dechlorinated to yield 1,2,4 trichlorobenzene. Since we found 1,2,4 trichlorobenzene inside Anabaena cells, another HCH dechlorinase or a similar, functionally equivalent enzyme must be catalyzing this reaction in this cyanobacterium.
In relation to Sphingomonas, although lin genes in Anabaena are just proposed, it is interesting to note the features concerning lin gene expression. In Sphingomonas, linB and linC are constitutively expressed whereas in Anabaena, the potential linB genes are constitutively expressed but the expression of the proposed linC gene is clearly inducible in the presence of lindane. Interestingly this linC induction was also observed in M. aeruginosa PCC 7806 cells (Sarasa-Buisán et al., 2022). The presence of lin genes and the inexistence of some metabolic intermediaries seem to suggest some alternative lindane metabolization pathway in Anabaena although an uncompleted pathway cannot be ruled out in which trichlorobenzene would be a dead-end product. Consequently, carefully designed experiments should be carried out in the future to clarify the lindane degradation pathway of Anabaena sp. PCC 7120.
In conclusion, the information provided in this work contributes to a better understanding of the physiological and transcriptional responses of Anabaena sp. PCC 7120 to lindane which would be helpful in the utilization of this cyanobacterium alone or taking part of consortia in the bioremediation of this pesticide. Anabaena sp. PCC7120 could be used in freshwater systems as the first step of lindane bioremediation to generate trichlorobenzene. Then, trichlorobenzenes can be reductively dechlorinated by organohalide-respiring bacteria in genera Dehalococcoides (Hölscher et al., 2003), Dehalobacter (Nelson et al., 2011), Dehalobium (Wu et al., 2002) and Dehalogenimonas (Qiao et al., 2018) or could be aerobically degraded by Proteobacteria such as Pseudomonas, Sphingomonas, and Xantobacter (Field & Sierra-Alvarez, 2007). Finally, it is interesting to point out that in this work, we also found a gene that is remarkably induced in the presence of lindane in Anabaena sp. PCC7120. This result is of great interest because this induction can be used in the development of sensing circuits for the generation of whole-cell biosensors based in the host Anabaena sp. PCC7120 for the detection of lindane.
AUTHOR CONTRIBUTIONS
Jorge Guío: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal). Maria F. Fillat: Funding acquisition (equal); supervision (equal). Maria L. Peleato: Conceptualization (equal); funding acquisition (equal); supervision (equal). Emma Sevilla: Conceptualization (equal); formal analysis (equal); supervision (equal); writing—original draft (lead); writing—review and editing (lead).
ACKNOWLEDGMENTS
We acknowledge the HCH analysis laboratories in Bailín (Sabiñánigo, Huesca) for the lindane analysis. Authors thank the financial support from Ministerio de Ciencia, Innovación y Universidades (grant PID2019-104889GB-I00), Gobierno de Aragón (grants E35_20R Biología Estructural).
CONFLICT OF INTEREST STATEMENT
None declared.
ETHICS STATEMENT
None required.
APPENDIX


| Primer | Sequence (5′–3′) | Gene |
|---|---|---|
| rnpB for | AGCGGAACTGGTAAAAGACCAA | Gene rnpB from Anabaena |
| rnpB rev | GAGAGGTACTGGCTCGGTAAACC | sp. PCC 7120 |
| linB1 for | CAGCTTTGGCGGCTCAAG | Gene all1353 from Anabaena |
| linB1 rev | GAAAAGCCAGAACCAATCCAAT | sp. PCC 7120 |
| linB2 for | CGATCGCACTCTCAAAGCTATAATC | Gene all0193 from Anabaena |
| linB2 rev | TCACATAGTAGCGCCAGATATGC | sp. PCC 7120 |
| linC for | GGAAAAGAAAGCAGTTGTAGAAAGTCA | Gene all3836 from Anabaena |
| linC rev | GCTACTGCTGCCGCCATTT | sp. PCC 7120 |
| linE for | ACTTCCGCATCTCTGCAAAAA | Gene all0352 from Anabaena |
| linE rev | CAGCCGTCTATTTGGCAACA | sp. PCC 7120 |
| linR for | AAATTCATGTTGCGTTGGGTTT | Gene alr0353 from |
| linR rev | CCAACAAAGTGTTCTTCAAATAATGG | Anabaena sp. PCC 7120 |
| sodA for | CTAACCAAACCCAACCACTACCA | Gene sodA from Anabaena |
| sodA rev | CCTTTGGCAGTTTTGAAGAGTTC | sp. PCC 7120 |
| catA for | ACCCACGGGATGTCATTATGA | Gene catA from Anabaena |
| catA rev | TGGCAACGCACCTAAACCA | sp. PCC 7120 |
Open Research
DATA AVAILABILITY STATEMENT
All data are provided in full in the results section of this paper and its appendix.