Highly expressed of SERPINA3 indicated poor prognosis and involved in immune suppression in glioma
Abstract
Introduction
The prognosis of patients with glioma is dismal. It has been reported that Serpin peptidase inhibitor clade A member 3 (SERPINA3) is associated with the mobility and invasion of tumor cells. Our study was designed to explore the value of SERPINA3 messenger RNA (mRNA) expression in the biological process, prognosis, and immune significance in glioma.
Methods
We analyzed the biological functions of SERPINA3 through data from the Chinese Glioma Genome Atlas databases. Differentially expressed genes and enrichment analysis were performed and correlations between SERPINA3 expression and immune cell infiltration were analyzed. Further, we validated the expression and the survival prediction role of SERPINA3 by using tissue microarrays and RNAscope in situ hybridization in 321 gliomas. The correlations between the expression and clinical-pathological parameters as well as other biomarkers were examined.
Results
Univariate and multivariate regression both indicated that the level of SERPINA3 transcript represented an independent prognostic factor. High levels of SERPINA3 correlated with poor survival in patients with glioma. Expression of SERPINA3 mRNA was observed positively correlated with MCM6, IGFBP2, and FKBP10. Enrichment analysis showed SERPINA3 mainly enriched in immune-related terms and signaling pathways including MAPK, TNF, P53, PI3K-Akt, nuclear factor-κB. Immune infiltration analysis further declare the SERPINA3 expression negatively correlated with levels of Macrophages M1, native CD4+ T cell, monocytes, and Mast cell activated. And overexpression of SERPINA3 correlated with low CD4+ T cell infiltration in glioma tissues.
Conclusions
SERPINA3 may play a key role in the biological process of glioma cells especially in immune suppression activities. SERPINA3 may serve as an independent survival prediction factor in glioma patients.
1 INTRODUCTION
Gliomas are the most prevalent primary tumours of the central nervous system, representing approximately 80% of malignant brain tumours1 and are responsible most deaths associated to primary brain tumours. Although concurrent radio- and chemoradiotherapy followed by chemotherapy after surgical removal of the tumour has become the standard treatment for patients with glioma, patient survival is still unsatisfactory. In particular, patients with high-grade glioma (World Health Organization grade IV) have a median overall survival (OS) of 15–17 months upon clinical management.2-4 In recent years, research has focused on aberrant molecular alterations in gliomas. IDH1/2 mutation,5 1p/19q co-deletion,6 TERT promoter mutation7 and several other markers have been used to define subtypes of gliomas. However, there are few therapeutic targets with potential clinical applications. Thus, more studies are needed to explore the specific molecular aberrations and propose new therapeutic regimens to improve the prognosis of patients.
Serpin peptidase inhibitor clade A member 3 (SERPINA3), also named alpha-1-antichymotrypsin, is a 55–66 kDa secreted serine protease that inhibits the activity of several serine proteases.8 Previous studies have shown that SERPINA3 plays an important role in cytokinesis, proliferation, apoptosis, and tumour metastasis.9, 10 Furthermore, SERPINA3 was reported to participate in multiple immunisation activities10 and that may serve as a potential immune therapy target in endometrial carcinoma.11 Nimbalkar et al.12 found that SERPINA3 contributes to proliferation, invasion, migration, and transition to mesenchymal phenotypes of glioma cells. Moreover, Luo et al.13 demonstrated that high levels of SERPINA3 messenger RNA (mRNA) are correlated with poor prognosis in patients with glioma through real-time polymerase chain reaction. However, SERPINA3 expression in glioma specimens detected by in situ methods has not been previously investigated, and the biological function of SERPINA3 in glioma has not been systematically analysed.
In the present study, the expression of SERPINA3 mRNA was first detected in 321 glioma and 13 normal tissues by RNA in situ hybridization. The relationship between SERPINA3 levels and the survival, clinical parameters, and expression of other biomarkers of glioma patients was analysed. Data from the Chinese Glioma Genome Atlas (CGGA) database were included as a test group. Furthermore, the biological function of SERPINA3 in glioma cells was explored. This study is expected to provide valuable information on the role of SERPINA3 as a novel biomarker and potential target in glioma.
2 MATERIALS AND METHODS
2.1 Data mining from public databases
Gene expression data and corresponding clinical information from the CGGA were downloaded (LGG + GBM) (http://www.cgga.org.cn/, DataSet ID: mRNAseq_693 and mRNAseq_325, Data type: RNA sequencing). Then we loaded these two sets of data into limma and sva packages in R software (R version 4.1.0: http://www.r-project.org/) to integrate and correction of gene expression data from glioma samples.
2.2 Differentially expressed genes (DEGs) and enrichment analysis
By analysis of gene expression data of CGGA, the sample of 1018 glioma patients were divided into two subgroups, according to the optimal cut-off point of SERPINA3 mRNA expression, SERPINA3high VS SERPINA3low. Gene expression differences between SERPINA3high and SERPINA3low were identified within DESeq. 2. DEGs were determined by Wilcoxon rank-sum test with q = 0.05 and fold change >1 after log2 transformation as the significance threshold and was used to create a volcano map. The genes with |log2Foldchange| >1.5 and adjusted p value of <.05 were regarded as the DEGs in the two groups. Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) on the DEGs between groups of SERPINA3high & SERPINA3low were performed by the clusterProfiler package. Adjusted p value of <.05 were seen as significant.
2.3 Analysis of immune cell infiltration
Correlations between tumor-immune infiltrating cells (TIICs) and the SERPINA3 expression were analyzed. The proportions of the 21 TIICs from each sample were determined by using the “CIBERSORT” (R package). Then the level correlation of any two of the 21 TIICs was cauculated through spearman test. Finally, for each of TIICs, the difference of immune cells containment was analyzed between subgroups of SERPINA3high and SERPINA3low.
To futher test the correlation of SERPINA3 expression with the immune inflammation of glioma, we analysis the immune inflammation through TIMER, CIBERSORT, CIBERSORT-ABS, QUANTI-SEQ, XCELL, and EPIC methods14 in glioma patients from The Cancer Genome Atlas (TCGA). Then correlation analysis between SERPINA3 mRNA expression and levels of immune cells were preformed through T test.
2.4 Tissue microarray (TMA) and in situ detection
A total of 321 gliomas and 13 paired normal or edematous brain tissue specimens were procured from the Department of Neurosurgery at the National Cancer Center/Cancer Hospital of Chinese Academy of Medical Sciences (CAMS). This research has been performed in accordance with the World Medical Association Declaration of Helsinki while approved by the Ethics Committee of CAMS (Number NCC2014G-12). The collection and preparation of glioma TMAs were performed as we previously done.15 RNAscope in situ hybridization (RISH) and immunohistochemistry (IHC) were used for in situ detection in glioma TMAs as previously described.15, 16 Following probes and antibodies were used in this study: SERPINA3 (NM_001085.4, bp 353–1345 ACD#412671); IGFBP2 (NM_000597.2, bp 490–1423, ACD#313061), while Ubiquitin C (UBC, NM_021009, bp 342–1503, ACD#310041) and DapB (ACD#310043) were used as positive and negative control. Anti-IDH1R132H antibody (working solution, ZM0447; ZSGB-BIO), anti-MCM6 antibody (1:100, 13347-2-AP; Proteintech), anti-FKBP10 antibody (1:2000, 50353; Sigma), and anti-CD4 antibody (working solution, ZA-0519; ZSGB-BIO) were used in IHC assay. The evaluation of in situ signal scores were performed as before.
2.5 Survival analysis
Survival and survminer packages were loaded in R. Surv_cutpoint function was used to determine the optimal cut-off point of SERPINA3 mRNA expression in the datasets of CAMS & CGGA, and therefore divided the samples into high and low SERPINA3 expression groups. OS curves were plotted according to the Kaplan–Meier method, with the log-rank test applied for comparison. Moreover, univariate along with multivariate Cox regression models were constructed to explore the role of SERPINA3 expression in survival of patients with glioma. Then time dependent receiver operator characteristics (ROC) curve analysis through survivalROC package in R was performed, at a threshold of area under curve (AUC) 0.7. Finally, a nomogram model including SERPINA3 mRNA level was established through rms package in R to predict the survival prognosis of glioma patients in 1, 2, and 3 years after the operation.
2.6 Statistical analysis
The wilcoxon rank-sum test was used to assess relations between the expression level of SERPINA3 and corresponding clinical information. Significant differences between two groups were determined by the Mann–Whitney U test. The χ2 test was used to assess the relationship between molecular alterations and clinico-pathological parameters. A p value of <.05 was considered statistically significant. All tests were two-sided.
3 RESULTS
3.1 Basic information of patients involved in the analysis
A total of 1339 patients with confirmed glioma (CAMS: 321 gliomas; CGGA:1018 gliomas) were collected the SERPINA3 mRNA expression data. However, after the exclusion of cases with incomplete survival information, the remaining cases (CAMS: 267 patients; CGGA:749 patients) were involved in the statistics. Baseline information of these patients could be found in Table 1.
| Variables | CAMS | CGGA | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | SERPINA3High | SERPINA3Low | p value | Total | SERPINA3High | SERPINA3Low | p value | |
| Gender | ||||||||
| Male | 149 | 23 (15.4%) | 126 (84.6%) | .296 | 442 | 244 (55.2%) | 198 (44.8%) | .059 |
| Female | 118 | 24 (20.3%) | 94 (79.7%) | 307 | 148 (48.2%) | 159 (51.8%) | ||
| Age (years) | ||||||||
| ≤50 | 125 | 13 (10.4%) | 112 (89.6%) | .004* | 542 | 250 (46.1%) | 292 (53.9%) | <.001* |
| >50 | 142 | 34 (23.9%) | 108 (76.1%) | 207 | 142 (68.6%) | 65 (31.4%) | ||
| Radiotherapy | ||||||||
| Yes | 157 | 21 (13.4%) | 136 (86.6%) | .331 | 625 | 328 (52.5%) | 297 (47.5%) | .86 |
| No | 90 | 17 (18.9%) | 73 (81.1%) | 124 | 64 (51.6%) | 60 (48.4%) | ||
| NA | 20 | 9 (45.0%) | 11 (55.0%) | |||||
| Chemotherapy | ||||||||
| Yes | 202 | 31 (15.3%) | 171 (84.7%) | .319 | 520 | 292 (56.2%) | 228 (43.8%) | .002* |
| No | 54 | 12 (22.2%) | 42 (77.8%) | 229 | 100 (43.7%) | 129 (56.3%) | ||
| NAa | 11 | 4 (36.4%) | 7 (63.6%) | 0 | ||||
| Gradeb | ||||||||
| WHO II | 3 | 0 | 3 (100.0%) | .047* | 218 | 59 (27.1%) | 159 (72.9%) | <.001* |
| WHO III | 7 | 0 | 7 (100.0%) | 240 | 109 (45.4%) | 131 (54.6%) | ||
| GBM | 257 | 47 (18.3%) | 210 (81.7%) | 291 | 224 (77.0%) | 67 (23.0%) | ||
| Pathology | ||||||||
| Primary | 231 | 36 (15.6%) | 195 (84.4%) | .013* | 502 | 236 (47.0%) | 266 (53.0%) | <.001* |
| Secondary | 0 | 0 | 0 | 25 | 20 (80.0%) | 5 (20.0%) | ||
| Recurrent | 3 | 0 | 3 (100.0%) | 222 | 136 (61.3%) | 86 (38.7%) | ||
| NA | 33 | 11 (33.3%) | 22 (66.7%) | |||||
| IDH mutation | ||||||||
| Mutant | 55 | 3 (5.5%) | 52 (94.5%) | .031* | 410 | 132 (32.2%) | 278 (67.8%) | <.001* |
| Wild type | 148 | 21 (14.2%) | 127 (85.8%) | 339 | 260 (76.7%) | 79 (23.3%) | ||
| NA | 64 | 23 (35.9%) | 41 (64.1%) | |||||
| living status | ||||||||
| live | 121 | 0 | 121 (100.0%) | .001* | 293 | 88 (30.0%) | 205 (70.0%) | <.001* |
| dead | 146 | 47 (32.2%) | 99 (67.8%) | 456 | 304 (66.7%) | 152 (33.3%) | ||
| Total | 267 | 47 (17.6%) | 220 (82.40%) | 749 | 392 (52.3%) | 357 (47.7%) | ||
- Note: Statistically significant difference (p value of <.05) and same below.
- Abbreviations: CAMS, Chinese Academy of Medical Sciences; CGGA, Chinese Glioma Genome Atlas; GBM: glioblastoma; WHO, World Health Organization.
- a NA, not available.
- b According to WHO 2016 classification.
3.2 Upregulated expression of SERPINA3 in glioma
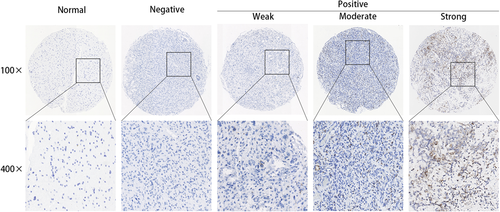
RNAscope in situ hybridization detection of CAMS sample showed the overexpression in glioma samples. All of the gliomas had positive RISH signals for UBC, but none had signals for DapB (Figure S1). Results of RISH showed that SERPINA3 was of high expression in 25.4% (68/267) of the tested gliomas. The positive signal is anatomically located in the cellular tumor area. Moreover, all the morphologically normal or edematous tissues presented negative SERPINA3 mRNA expression (Figure 1).
3.3 SERPINA3 mRNA level correlated with clinical characteristic
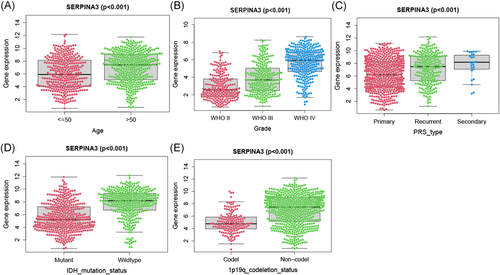
We examined the relationship between SERPINA3 expression level and clinic-pathological features of patients in CAMS and CGGA data (Table 1). The results showed that SERPINA3 was significantly higher expressed in GBM than that in LGG (p = .047 and p < .001 in CAMS and CGGA, respectively. And same below), and in primary glioma than secondary (p = .013 and p < .001). Elderly patients (>50 years) tended to present with a higher level of SERPINA3 mRNA expression than younger patients (≤50 years) (p = .004 and p < .001). Besides, the expression level of SERPINA3 showed a strong correlation with IDH1 mutation and 1p19q co-deletion status (p < .001, Table 1 and Figure 2).
3.4 SERPINA3 mRNA expression associated with other biomarkers of gliomas
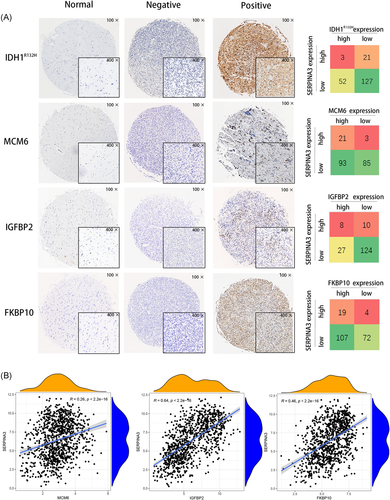
For the same cases, we previously reported high expression of IDH1 mutation,17 MCM6 protein,16 FKBP10 protein,18 and IGFBP2 mRNA.15 Here, we simultaneously analyzed the relationship between SERPINA3 expression and the alterations biomarkers. We found that levels of SERPINA3 was positively correlated with the expression of MCM6 protein, IGFBP2 mRNA, and FKBP10 protein (p = .001, .014, and .039, respectively, Table 2 and Figure 3A). Further, we examined the expression data of SERPINA3 and these biomarkers in CGGA datasets and the transcript level of SERPINA3 showed positively correlated with MCM6, IGFBP2, and FKBP10 (Figure 3B).
| Biomarker | Status | SERPINA3 | χ2a | p value | |
|---|---|---|---|---|---|
| High | Low | ||||
| IDH1 | Mutation | 3 | 21 | 4.652 | .031* |
| Wild type | 52 | 127 | |||
| MCM6 protein | High | 21 | 3 | 9.304 | .001* |
| Low | 93 | 85 | |||
| IGFBP2 mRNA | High | 8 | 10 | 6.911 | .014* |
| Low | 27 | 124 | |||
| FKBP10 protein | High | 19 | 4 | 3.607 | .039* |
| Low | 107 | 72 | |||
- Note: Statistically significant difference (p value of <.05) and same below.
- Abbreviations: CAMS, Chinese Academy of Medical Sciences; FKBP10, FK506 binding protein 10; IDH1, isocitrate dehydrogenase 1; IGFBP2, insulin-like growth factor binding protein 2; MCM6, minichromosome maintenance 6; mRNA, messenger RNA; SERPINA3, Serpin peptidase inhibitor clade A member 3
- a Chi-square test of the expression of SERPINA3 and other biomarkers.
3.5 SERPINA3 serves as a prognostic biomarker in gliomas
Since the 97.0% (259/267) of patients in CAMS data are GBM, we could only observed the prognosis value of SERPINA3 in GBM. Kaplan–Meier survival analysis showed that the expression levels of SERPINA3 were negatively related to the prognosis of patients with GBM (n = 259, p < .001, Figure 4A). Subsequently, the prognosis value of SERPINA3 mRNA was validated in the data of CGGA through the same methods. The results indicated that the high level of SERPINA3 mRNA showed a strong correlation with the dismal prognosis of patients in both LGG and GBM (n = 291 and 458, respectively, both p < .001, Figure 4B,C).
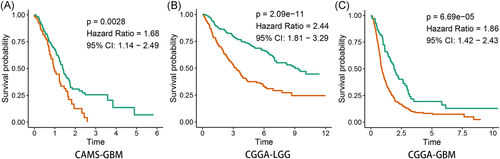
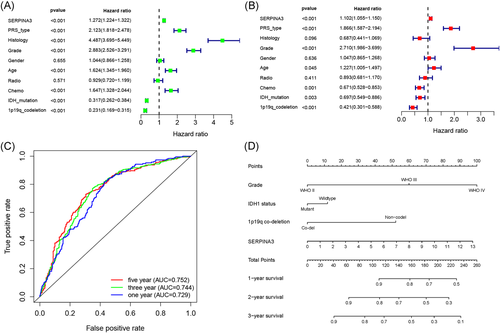
Result of univariate and multivariate Cox analyse indicated that high expression of SERPINA3 was an indepent shorter survival indicator for glioma patients (Figure 5A,B). Tine-ROC analyses showed that AUC of SERPINA3 expression in 1, 3, 5 years was 0.752, 0.744, 0.729, respectively (Figure 5C). Combining with clinical parameters including grade, IDH1 mutation, and 1p19q co-deletion status, we could predict the survival time of glioma patients more precisely. The results were presented in the nomogram (Figure 5D) and had been validated (shown in Figure S2).
3.6 DEGs and enrichment analysis
We compared the gene expression differences between high and low SERPINA3 expression groups in CGGA using the DESeq. 2 package. A total of 3280 genes and 829 genes were identified as upregulated and downregulated DEGs, respectively. The DEGs were exhibited in the volcano map (Figure 6B). The expression of the 20 genes with the largest differences in up- and downregulation in CGGA individuals was presented in the heatmap (Figure 6A). The correlation between any two of the 40 genes was shown in Figure 6C.
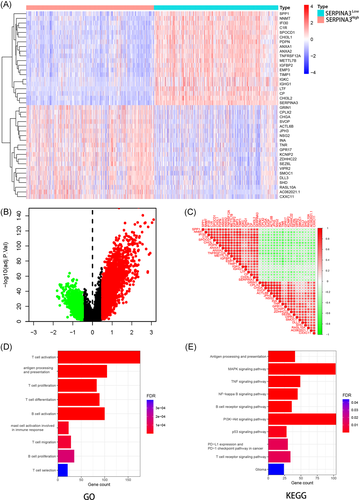
GO enrichment analysis showed that SERPINA3 were enrichment in various immune-related terms including antigen processing and presentation, T and B cell activation, differentiation, and proliferation (Figure 6D). KEGG pathway analysis showed that SERPINA3 were mainly enriched in MAPK, TNF, P53, PI3K-Akt, nuclear factor-κB signaling pathway and PD-L1 expression and PD-1 checkpoint pathway in cancer (Figure 6E).
3.7 SERPINA3 expression correlated with immune infiltration level and TME
We further analyzed the relationship between SERPINA3 expression and immune cells infiltrating tumors. The comparative content of immune cells in each case was represented by a barplot (Figure 7A) and correlations among various clusters of immune cells were showed (Figure 7B). We invested the differences of in the immune cells level between subgroups SERPINA3high VS SERPINA3low. The results showed that high expression of SERPINA3 was negatively correlated with levels of M1 Macrophages, CD4 native T cell, monocytes, and Mast cell activated (p = .017, .013, .025, and .018, respectively, Figure 7C). Immune infiltration analysis of TCGA glioma patients in different methods showed similarty results (Table S1).
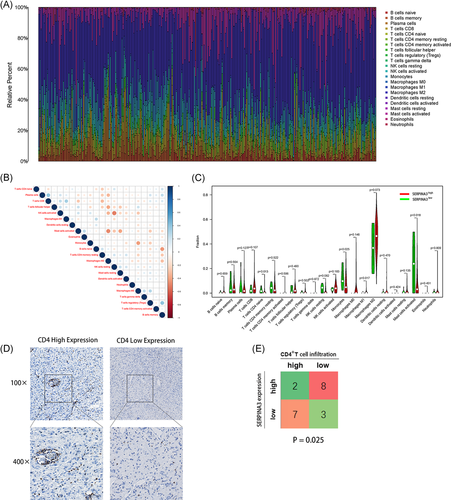
CD4+ T cell play a vital part in the immune procession of glioma.19 To further test the correlation between SERPINA3 mRNA expression and the tumor immune infiltrating of CD4+ T cell in tissues, we performed the IHC experiment in 20 gliomas. The density of CD4 signal were divided into high- and low-score groups separately according to the IHC staining (Figure 7D). As shown in Figure 7E, 8/10 SERPINA3high patients showed low CD4+ T cell in-filtration while 3/10 SERPINA3low patients showed low CD4+ T cell infiltration (p = .025).
4 DISCUSSION
Gliomas are one of the most aggressive and common cancers, therefore, exploring novel molecular alterations in glioma may provide valuable insights for the design of effective therapeutic strategies. This study showed that SERPINA3 expression is associated with glioma malignancy, including higher grade tumours and shorter patient survival time. Moreover, SERPINA3 may play a crucial role in the immune response. Specifically, SERPINA3 was found to be negatively associated with M1 Macrophages, T cell CD4 native, monocytes, and activated mast cells. Moreover, expression of SERPINA3 correlated with low CD4+ T cell infiltration in glioma tissues. Overexpression of SERPINA3 mRNA in high grade gliomas suggests that SERPINA3 may play a key role in the progression of glioma cells.
Herein, the proportion of SERPINA3 overexpression in glioma tissues ranged from 17.6%–52.3% in different datasets. In addition to the variation between the CGGA and CAMS samples, one of the reasons for such a large difference may be due to different detection methods. In the CGGA dataset, SERPINA3 was measured by transcriptome sequencing whereas RNAscope in situ hybridisation, which has high specificity and sensitivity, thereby providing more visible and accurate results, was used to evaluate the CAMS samples. In addition, the threshold of high expression was determined based on the prognostic analysis of various datasets, with glioma patients with high SERPINA3 expression tending to have poor survival. In agreement with this finding, Luo et al.13 reported that expression of SERPINA3 was correlated with poor survival in glioma patients, as evaluated by immunohistochemistry. Recent studies have shown that SERPINA3 acts as a key molecule for survival prediction in various cancers.11, 20 The present study not only demonstrates that SERPINA3 is a negative prognostic indicator, but also that it has a valuable prognostic performance for 1-, 3-, and 5-year survival. Moreover, a survival prediction model based on SERPINA3 mRNA expression and other factors in glioma patients was established for the first time. These analyses showed that SERPINA3 can play a significant role in predicting the survival of glioma patients.
A previous study found that SERPINA3 promotes the proliferation of melanoma and human liver cancer cells.20, 21 In the present study, the relationship between SERPINA3 and other previously verified proliferation-related molecules, including MCM6,16IGFBP2,15 and FKBP1018 was evaluated. A strong correlation between the expression of SERPINA3 and these molecules in glioma suggests that SERPINA3 may participate in the proliferation of glioma cells. In addition, SERPINA3 was related to the PI3K/Akt and MAPK/Ras/Raf pathways, as indicated by the biological function analysis. These pathways have been shown to be involved in the regulation of malignant glioma cell proliferation.22, 23 Taken together, these results indicate that SERPINA3 may promote the proliferation and strengthen the malignant progression of glioma cells.
In the present study, biological analysis showed that SERPINA3 is highly enriched in multiple immune-related terms, including activation, differentiation, and proliferation of T cells, which was in accordance with a previous study on endometrial carcinoma.11 Thus, it is reasonable to assume that SERPINA3 may play an essential role in the immune process of cancer. Increasing evidences have demonstrated the importance of the tumour microenvironment in cancer development, and an increasing number of studies have shown that tumour-infiltrating immune cells can serve as promising indicators of therapeutic effects.24 Infiltrated inflammatory cells, tissue-resident cells, and brain vasculature constitute the glioma microenvironment.25 Moreover, a reduced number of inflammatory cells in the glioma microenvironment may prevent an efficient immune surveillance.26 Herein, the role of SERPINA3 in the regulation of the tumour microenvironment in glioma tissues was analysed in detail for the first time. Crosstalk between microglia and peripheral macrophages in the microenvironment of glioma can block the immune response of effector T cells through the activation of immune checkpoint proteins, such as B7-H4.27 Macrophage differentiation is induced and regulated by various microenvironmental signals, leading to M1, which can promote inflammatory reactions, and M2, which can regulate immunity, repair, and remodelling tissue. Given that SERPINA3 levels were herein negatively correlated with the number of M1 macrophages, suggests that SERPINA3 may downregulate the inflammatory reaction of glioma cells.
A recent study showed that SERPINA3 is a novel target of STAT3.21 Activation of STAT3 is critical for tumour-induced immune tolerance and avoidance in the glioblastoma microenvironment. For example, interleukin-2-mediated STAT3 activity expands tumour-associated regulatory T cells and enhances Foxp3 expression in CD4+ T cells.28 Based on these findings, whether SERPINA3 affects CD4+ T cells in glioma by affecting STAT3 should be further investigated. Alternatively, monocytes are important regulators in cancer development and contribute to antitumour immunity through phagocytosis, remodelling of the extracellular matrix, and recruitment of lymphocytes.29 We found that the number of monocytes in the tumour intra-environment was reduced in the SERPINA3high group, which may be a possible method for the regulation of immunity. These results suggest that SERPINA3 is involved in promoting immunosuppression in the glioma microenvironment.
There is now high hope for the efficacy of various immune therapy strategies in glioma based on immune checkpoint inhibition. Anti-PD-L1 antibodies, such as nivolumab or pembrolizumab, have shown powerful antitumour effects. The relationship between PD-L1 and T cell infiltration in glioma, as well as the therapeutic effects of anti-PD-1/PD-L1 antibodies, remain largely elusive, which may reflect the specificity of the cellular and structural microenvironment in the brain.30 Multiple regulators influence PD-L1 expression at the RNA and protein levels and drug intervention via direct regulation.31 The present analyses showed that high SERPINA3 expression was positively correlated with PD-L1 expression (p < .001), suggesting that SERPINA3 may be involved in the PD-L1-mediated immune escape pathway. Further experiments should be performed to explore the inner mechanisms between these two molecules, and the therapeutic effect of combined inhibitory approaches should be validated.
5 CONCLUSION
The present study demonstrates that SERPINA3 is overexpressed in glioma tissues and is involved in the proliferation of glioma cells. Moreover, SERPINA3 promotes immune suppression in the glioma microenvironment. Thus, SERPINA3 may serve as a novel prognostic biomarker and therapeutic target in glioma. Nonetheless, analysis of large-scale dataset and mechanistic studies are still needed to validate the function of SERPINA3 in glioma.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (82072803).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Jing-Hai Wan and Ming-Rong Wang designed the study, analyzed the data, and revised the manuscript. Qing Yuan designed the study, performed the experiments, analyzed the data and drafted the manuscript. Hong-Qing Cai designed the study, analyzed the data, and revised the manuscript. Song-Quan Wang, Guang-Tao Zhang, Zhi-Dan Liu, and Jie He contributed materials, collected clinical information, performed the experiments and analyzed the data.
Open Research
DATA AVAILABILITY STATEMENT
Data of CGGA part during the study are available in a repository or online in accordance with funder data retention policies (http://www.cgga.org.cn/).
Data of CAMS part during the study are available from the corresponding author by request ([email protected] [J-H.W]; and [email protected] [H-Q.C]).