scICB: A pan-cancer database of human temporal immune checkpoint blockade therapy at single-cell transcriptomic resolution
Fansen Ji, Weitong Bi and Jiawei Zhang are joint authors
Dear Editor,
We developed a pan-cancer scRNA-seq database under the treatment of Immune checkpoint blockade (ICB): scICB. The detailed biopsy timepoint relative to the ICB and clinical efficacy assessment information after the treatment have been identified to help clinicians to analyse different aspects of immunotherapy responsive biomarkers (Figure 1). The database is freely accessible at http://www.scimmnue.com/.


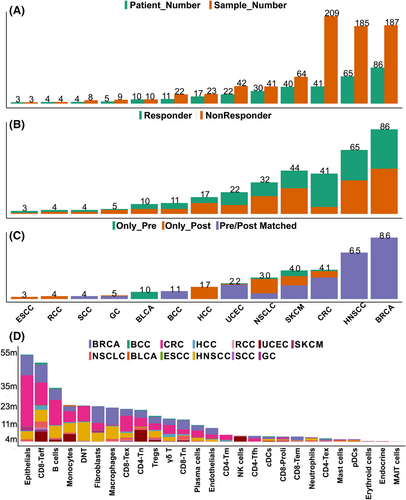
Herein, we collected scRNA-seq datasets related to ICB treatment and corresponding clinical information from multiple sources (Table 1). scICB includes 807 samples from 338 patients treated by ICB or ICB combination therapy across 13 cancer types (Figure 2a, Tables S1–S3). A total of 3 686 385 single cells covering NSCLC, CRC, RCC, HNSCC, BLCA, BCC, SCC, HCC, BRCA, SKCM, ESCC, GC and UCEC. For response status level, there are 174 patients defined as responders (R) while 170 patients labelled as nonresponders (NR) (Figure 2b). For biopsy timepoint level, a total of 23 patients only has pre ICB treatment scRNA-seq data, while a total of 68 patients only has post ICB treatment data and a total of 247 patients has matched pre and post treatment scRNA-seq data (Figure 2c), which facilitate us to trace the dynamic TME changes during the intervention of ICB. After annotating the major cell type and removing mitochondrial or ribosome genes enriched cells, we have annotated the cell types for each cell (Figure 2d). BRCA, HNSCC, CRC, SKCM and NSCLC rank top 5 both in terms of both patient and sample number in scICB, implying the high level of clinical translational research activity for ICB in these cancer types over the past few years.
| ID | Cell number | Cancer type |
|---|---|---|
| 1 | 289 046 | (CRC, Colorectal Cancer) |
| 2 | 80 513 | (NSCLC, Non-Small Cell Lung Cancer) |
| 3 | 32 399 | (HNSCC, Head and Neck Squamous Cell Carcinoma) |
| 4 | 19 053 | (RCC, Renal cell carcinoma) |
| 5 | 87 589 | (NSCLC, Non Small Cell Lung Cancer) |
| 6 | 11 307 | (BLCA, Bladder Carcinoma) |
| 7 | 28 081 | (NSCLC, Non Small Cell Lung Cancer) |
| 8 | 53 030 | (BCC, Basal Cell Carcinoma) |
| 9 | 26 016 | (SCC, Squamous Cell Carcinoma) |
| 10 | 175 942 | (BRCA, Breast Invasive Carcinoma) |
| 11 | 50 693 | (BRCA, Breast Invasive Carcinoma) |
| 12 | 217 275 | (BRCA, Breast Invasive Carcinoma) |
| 13 | 83 793 | (HCC, Hepatocellular Carcinoma) |
| 14 | 16 291 | (SKCM, Skin Cutaneous Melanoma) |
| 15 | 326 056 | (HNSCC, Head and Neck Squamous Cell Carcinoma) |
| 16 | 120 324 | (HNSCC, Head and Neck Squamous Cell Carcinoma) |
| 17 | 29 684 | (HCC, Hepatocellular Carcinoma) |
| 18 | 214 914 | (HNSCC, Head and Neck Squamous Cell Carcinoma) |
| 19 | 12 881 | (ESCC, Esophageal Squamous Cell Carcinoma) |
| 20 | 252 249 | (UCEC, Uterine Corpus Endometrial Carcinoma) |
| 21 | 15 069 | (SKCM, Skin Cutaneous Melanoma) |
| 22 | 158 925 | (BRCA, Breast Invasive Carcinoma) |
| 23 | 193 873 | (BRCA, Breast Invasive Carcinoma) |
| 24 | 161 697 | (BRCA, Breast Invasive Carcinoma) |
| 25 | 376 033 | (CRC, Colorectal Cancer) |
| 26 | 270 480 | (CRC, Colorectal Cancer) |
| 27 | 327 935 | (CRC, Colorectal Cancer) |
| 28 | 55 237 | (GC, Gastric Cancer) |
scICB provides mainly four functionalities for users. In Browse module, users can browse information like Cancer Type, Dataset ID, Tissue Type, Patient ID, Sample ID, Cell Type, Timepoint, Response and ICB Drug. Besides, the logFC value expression table for each cell type, TNSE, UMAP, marker gene heatmap plots, cell type annotation and relative proportion for each patient, patient/response status/biopsy timepoint/tissue type can also be browsed (Figure S1). In Pre VS Post module, users could select an interested cancer type and dataset to see the dynamic changes before and after the ICB for a certain tissue type and cell type and the relevant volcano plot, differential genes and enrichment analysis will be returned (Figure S2). In R VS NR module, users can compare the DEGs between R and NR for a certain cell type after selecting a certain cancer type and dataset ID, helping to uncover the underlying mechanisms or biomarkers of immune response to ICB (Figure S3). In GeneSet module, we provide functionality for users to upload custom gene sets or specific signalling pathway genes for analysis using our curated datasets. After selecting the cancer type, dataset, tissue type, and cell type, and uploading their gene sets, users will receive a boxplot illustrating gene set activity across different timepoint-response combinations (Figure S4).
In our pan-cancer level integrative analysis, we first investigated DEGs across diverse cell types in response to ICB treatment. When comparing Pre and Post ICB treatment, as well as Responders (R) versus Nonresponders (NR), we observed considerable variability in the number of DEGs across different cell types (Figures 3a and S5), reflecting the inherent heterogeneity of the datasets. We then conducted a cell type enrichment analysis across datasets containing different tissue types using the Ro/e method.1 In tumour tissues, we found endothelial cells, mast cells, CD8 exhausted T cells, plasma cells, and macrophages were significantly enriched, indicating biological process such as angiogenesis, antigen presentation, T cell cytotoxicity and humoral immunity is upregulated in tumour tissues.2-4 Fibroblasts and epithelial cells were enriched not only in tumour tissues but also in normal tissues, suggesting their involvement in both malignant and non-malignant environments.5 In peripheral blood mononuclear cells (PBMCs), monocytes, double-negative T cells (DNT), CD8 effector T cells, and γδ T cells were predominantly enriched, indicating their circulating nature in immune response to cancer.6-8 On the other hand, B cells, a major component of lymph nodes, were enriched in tumour-draining lymph nodes (tdLNs, Figure 3b), consistent with previous studies that B cells can form germinal centres in tdLNs and may serve as a type of antigen presenting cells for immune response.9 These results highlight distinct immune microenvironments across tissue types.
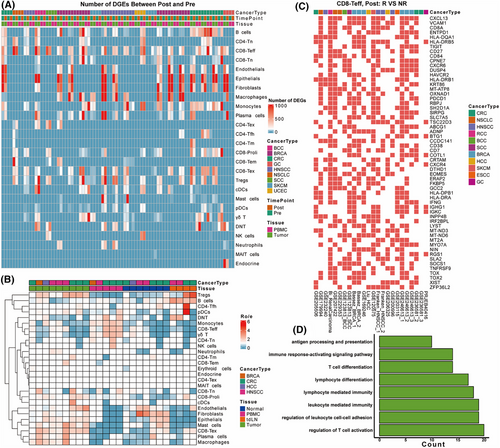
Given the pivotal role of CD8 effector T cells (Teff) in antitumour immunity, we analysed DEGs between R and NR in this cell type, specifically within tumour tissues after ICB treatment (Post). CD8 effector T cells were identified in 25 out of 28 datasets. We ranked genes by the number of datasets in which they were differentially expressed and select those found in more than 10 datasets and termed these genes as T cell responsive gene set (TRS) (Figure 3c). Among TRS, CXCL13 was upregulated in 18 out of 28 datasets. Other highly expressed genes in responders included VCAM1 (14/28 datasets), CD8A (12/28 datasets), and ENTPD1 (12/28 datasets). Immune checkpoint molecules were also significantly upregulated in responders. TIGIT (12/28 datasets), HAVCR2 (11/28 datasets, also known as TIM-3), and PDCD1 (11/28 datasets, encoding PD-1) were among the top immune checkpoint genes. The upregulation of these inhibitory molecules may be targeted by ICB and activate the T cell cytotoxicity. Further Gene Ontology (GO) enrichment analysis on the DEGs differentially expressed in more than 10 datasets revealed significant enrichment of processes related to T cell activation, differentiation and antigen presentation (Figure 3d). In summary, our results underscore the complex interplay of immune cell types and gene expression patterns in determining patient responses to ICB therapy. The identification of key DEGs and enriched biological processes provides valuable insights into the mechanisms driving successful immune responses in cancer immunotherapy.
In conclusion, we built scICB, a pan-cancer database of scRNA-seq database under the treatment of ICB. Using T cells in scICB as an example, we analysed the T cell responsive biomarkers that are predictive to ICB efficacy, which can facilitate the clinicians and researchers to explore their own biomarkers interested. More Information can be seen in Supplementary Notes.
AUTHOR CONTRIBUTIONS
Fansen Ji: Data curation; methodology; validation; formal analysis; investigation; writing—original draft. Weitong Bi: Formal analysis; methodology. Jiawei Zhang: Data curation; investigation. Bingjun Tang: Resources. Ying Xiao: Resources. Huan Li: Resources. Hao Liu: Resources. Boyang Wu: Resources. Fei Yu: Resources. Shizhong Yang: Conceptualisation; writing—review & editing; supervision; Project administration. Gang Xu: Conceptualisation; methodology; writing—review & editing. Jiahong Dong: Conceptualisation; resources; writing—review & editing; supervision; project administration; funding acquisition. All authors have read and approved the final manuscript.
ACKNOWLEDGEMENTS
All contributors to this work have been listed as authors in the manuscript. Therefore, no additional acknowledgments are necessary.
CONFLICT OF INTEREST STATEMENT
The authors have declared no conflict of interests.
FUNDING INFORMATION
This work was supported by the Natural Science Foundation of China (Grant Nos. 82090052, 82090050, and 82090053), and Tsinghua University Initiative Scientific Research Program of Precision Medicine (2022ZLA007).
ETHICS STATEMENT
This study does not involve in-house human participants, animal experiments, or clinical data requiring ethical approval.
Open Research
DATA AVAILABILITY STATEMENT
All scRNA-seq datasets used in this paper can be browsed and downloaded as Seurat files on the scICB database: http://www.scimmnue.com/.




