Inflammatory-associated myeloid dendritic cells reveals associations between chronic lung diseases and lung cancer
ABSTRACT
The high risk of patients with chronic lung diseases in developing into lung cancer has been recognised, but the key factors driving such procedure are still barely known. Dendritic cells (DCs) as major antigen presenting cells take part in the immune response in the very upstream, and myeloid DCs regulate the inflammation in pulmonary diseases. In this article, we performed single-cell RNA sequencing (scRNA-seq) analyses on DCs from pulmonary diseases. We explore the DC characteristics in chronic lung diseases, lung cancer and healthy control samples. We discover that a special type of DC, which is highly associated with inflammatory, inf-DC, is abundant in lung cancer samples. Furthermore, we find that there are about 10% patients with chronic lung diseases also has such inf-DC-rich pattern. Such proportion is consistent to the fact that about 10% chronic lung disease patients finally developed into cancer. Our findings indicate inf-DC could be a potential factor to predict the risk of chronic lung disease developing into cancer.
1 INTRODUCTION
In recent decades, the incidence of lung diseases including idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD) and lung cancer has rapidly increased and become a major threat to human life, which might be caused by unhealthy lifestyles, such as tobacco smoking.1-3 There is evidence that chronic lung diseases such as IPF and COPD are closely associated with lung cancer4-6; however, the relationship between these chronic lung diseases and lung cancer has not been fully understood. These pulmonary diseases share common properties with lung cancer in pathogenesis and predispositions such as immune dysfunction, and patients with IPF and COPD are at high risk of developing into cancer.7, 8 Recent technological advance in scRNA-seqopens up an alternative approach to understand pulmonary diseases,9-11 and opportunity to study the pathological mechanism between different pulmonary diseases.
Dendritic cells (DCs) play an important role in immune response as the major antigen presenting cells and in the induction of anti-tumour T-cell immunity.12 It has been known that myeloid DC participates in the inflammation regulation,13, 14 which is a key pathological feature of IPF, COPD, lung adenocarcinoma (LUAD), etc. In this article, we re-analysed scRNA-seq data collected from multiple pulmonary diseases. Within these datasets, three myeloid DC subtypes were identified: type 1 conventional DC (cDC1), type 2 conventional DC (cDC2) and mature DC (mature-DC). Each of the three subtypes could be further divided into two subsets, which were annotated as inflammatory DC (inf-DC) and non-inflammatory DC (non-inf-DC) based on their expression features of IRF8 and pro-inflammatory cytokines and chemokines.15 We found a specifically high abundance of inf-DCs in lung cancer patients (100%), while non-inf-DCs were the majority in normal tissue. Critically, we also found that about a certain proportion of IPF and COPD patients were dominated by inf-DCs, which shared the same pattern with lung cancer patients, such as LUAD. The proportion of such patients in IPF and COPD was very close to the reported proportion of patients of these chronic diseases developing into lung cancer.16, 17 Besides, the distant normal tissues from cancer patients were considered as normal control in the original study,18 and showed the same inf-DCs abundance pattern (100%) as LUAD tissues. We further investigated the cell‒cell interaction between different subtypes of DCs and lymphocyte cells. The results indicated that inf-DCs were more active in innating lymphocyte cells through ICAM-119; and such pattern was shared between IPF and lung cancer, but not found in COPD. The inf-DCs interacted with other cells differently in various lung diseases, but they shared the same pathogenic genes such as SEMA7A.20, 21 It appeared in DCs of all lung chronic diseases and lung cancer tissue, but not in normal samples.
In this article, we propose that the inf-DC in chronic lung diseases such as IPF and COPD share a similar pattern with that of lung cancer, which indicates their pathogenic similarity. And the inf-DC pattern in patients with chronic lung diseases could be the potential prediction factor for the risk of early lung cancer onset. We noticed that there existed one predictive model based on a series of inflammatory markers for COPD cases.22 Here, we propose that in the scale of single cell, the significant inf-DC abundant (0 or 1) pattern along with its markers in peripheral blood mononuclear cell (PBMC) could be a more effective indicator.
2 RESULTS
In this study, we analysed scRNA-seq datasets with tissues from several common lung diseases. The data collected are detailed in Table 1.
We collected a total of 15 788 DCs from myeloid cells from the datasets (Table 1), including healthy control (normal, NOR), IPF, COPD, systemic sclerosis (SSC), para-cancer (PC-NOR), LUAD, large cell carcinoma (LCC), and applied Seurat pipeline analysis. We finally included 43 NOR23 samples (seven distant normal tissue separated from the malignant region by at least 5 cm from LUAD patients18), 37 IPF23 samples, 16 COPD23 samples, seven SSC24 samples, five PC-NOR samples, 19 LUAD25 samples and three LCC25 samples. We integrated the data and further categorised these cells into three subtypes, that is, cDC1, cDC2 and mature-DC by canonical marker genes (Figure 1a‒d).26 The distribution of DC subtypes also demonstrated disease specific patterns. DCs from the tissues of cancer patients (PC-NOR, LUAD and LCC) only appeared in clusters on the right side (Figure 1c, clusters 1, 6, 8, 10, 12 and 13). DCs from normal tissues of healthy donors, as well as from patients with chronic lung disease, IPF and COPD, were present on all clusters, while DC from SSC patients only appeared on the left clusters. cDC2 was the most abundant subtype in each of the lung diseases, which counted for more than 70% of all DC cells. We also discovered the DC subtype composition of each pulmonary disease (Figure 1e), and discovered that the total number of DCs in cancer (LUAD, LCC and PC-NOR) was lower than that of normal patients and chronic diseases. The percentage of cDC2 in LUAD and PC-NOR was significantly higher than that of NOR and IPF, while the percentage of cDC1 was significantly lower (Figure 1f), a pattern consistent with previous reports.27, 28 The results indicated that DC in cancer tissues showed a distinct clustering pattern and DC subtypes compositions, while that of chronic diseases except for SSC were similar with normal sample.
| Disease | Dataset | Number of patients included |
|---|---|---|
| NOR | GSE136831,23 GSE128169,24 GSE12803341 | 36 |
| Distant NOR form LUAD patients | GSE131907_Lung_Cancer18 | 7 |
| COPD | GSE136831 | 16 |
| IPF | GSE136831, GSE128033 | 37 |
| SSC | GSE128169 | 7 |
| PC-NOR | E_MTAB_6653,25 E-MTAB-614925 | 5 |
- Abbreviations: COPD, chronic obstructive pulmonary disease; IPF, idiopathic pulmonary fibrosis; LUAD, lung adenocarcinoma; PC-NOR, para-cancer normal; SSC, systemic sclerosis.
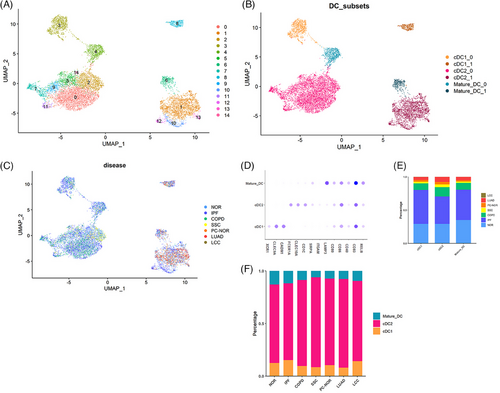
2.1 Disease associated subset division in each of the three DC subtypes
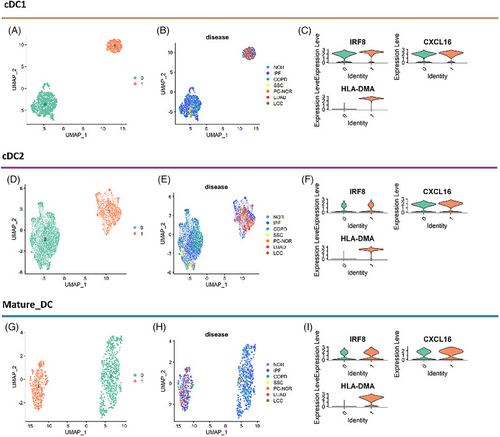
The DCs from different lung diseases have consistent clustering patterns, suggesting that there might be similar functional subsets of DCs in different lung diseases. Thus, we explored the relationship between these diseases in each of the three DC subtypes. As exhibited on the UMAP, each of the three DC subtypes showed similar patterns related to diseases (Figure 2a‒g). Each of the DC subtypes was clustered into two subsets (clusters 0 and 1). The tissues from normal control, IPF patients and COPD patients contained both subsets of DCs, while SSC tissue only contained cluster 0, and lung cancer tissue including para-cancer only contained cluster 1. We found higher gene expression of pro-inflammatory cytokines and chemokines, and IRF8 (Figures 2c‒i and 3c) in cluster 1 subset in every DC subtype (particularly in cDC2), and identified them as inf-DC.15
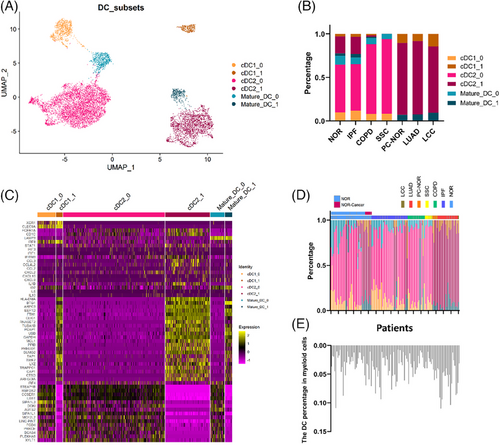
2.2 The inf-DC subset in patients with chronic lung diseases indicates risk of developing cancer
We divided DCs into six subtypes: cDC1_0, cDC1_1, cDC2_0, cDC2_1, mature-DC_1 and mature-DC_0. In fact, we first found that cDC2_1 showed a higher expression of genes related to type I interferon (IFN) (STAT1, IFIT1) and pro-inflammatory cytokines (CCL3, IL1B) as reported,15 and then confirmed its higher expression IRF8 (Figure 3c). The same pattern was observed in cDC1_1 and mature-DC_1. Besides, we also applied a new algorithm scSTAR29 to compare the two subsets of DC, which confirmed the gene expression characteristics of inf-DC on the cluster 1 subset more clearly (Figure S1).
Therefore, we annotated the cluster 1 subset of each DC subtype as inf-DC, and the other as non-inf-DC. We also identified the common marker genes between inf-DC and non-inf DC, and the markers of inf-DC generally promoted the power of antigen presenting30 (Figure S2). Notably, LUAD, LCC and PC-NOR only contained inf-DCs, while in other cases, non-inf DC took the major part (Figure 3b). By analysing the DC distribution of each patient, we found that the percentage of inf-DC was particularly patient related. For most patients, the percentage of inf-DC was either 0 or 1. We observed that the percentages of inf-DC in patients from cancer were all 100%, and the percentage of DCs in myeloid cells were higher than that of other lung diseases (Figure 3d,e). A part of the normal tissues and IPF samples had the same pattern with cancer, that is, extremely higher proportion of inf-DC and higher proportion of DC in myeloid cells (Figure 3d,e). More detailly, 8.33% of normal control, 13.5% of IPF patients and 12.51% of COPD patients showed high inf-DC pattern. We noticed that patients with IPF was reported to have about 13.54% probability of developing lung cancer,16 and that of COPD was about 8.8%.17 Furthermore, we found that 100% of the NOR-Cancer (distant normal tissue from LUAD patients) has this inf-DC pattern. Based on this, we suppose inf-DC composition features is associated with cancer developing in chronic lung diseases.
2.3 The inf-DC acquired a higher ability to activate lymphocyte cells, and shared similar pathogenic genes in IPF, COPD and LUAD
Comparing inf-DC with non-inf DC, we utilised CellPhoneDB31 to infer cell‒cell communications between DCs and lymphocytes based on the relative abundance of ligand‒receptor (L‒R) pairs, and performed a hallmark analysis. We found that cDC2 interacted more actively with lymphocytes than cDC1 and mature-DC; and inf-DC were generally more active than non-inf-DC (Figure 4a‒c). Besides, the overall DC‒lymphocyte interaction in IPF and LUAD was much more active than that in other diseases. We further compared inf-DC to non-inf DC in each of the three DC subtypes and performed gene set enrichment analysis (GSEA) hallmark analysis.32 All three inf-DC gene sets are enriched MYC_TARGETS_V1, which is related to genomic instability, cell differentiation and poor survival.33 Besides, they are enriched Interferon_Gamma_Response, and their main functions are inflammation and regulation of apoptosis. And previous research pointed out that activated MYC signalling showed higher IFN-γ response.33 However, the up-regulated DE genes in inf-cDC2 are enriched in INTERFERON_ALPHA_RESPONSE, indicating its ability of promoting cancer immunity.34, 35
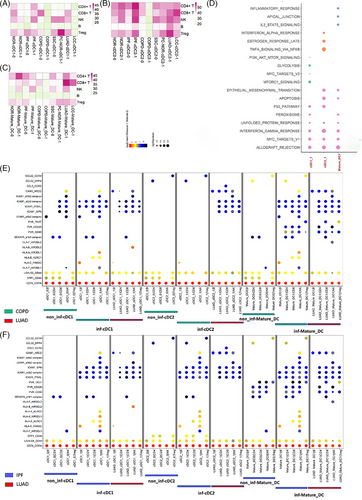
Based on the relative abundance of receptor-ligand pairs, we compared the cell‒cell interaction features of inf-DC and non-inf DC between chronic lung disease and LUAD (Figures 4e,f and S3,S4). Generally, all of the inf-DCs in IPF and LUAD indicated more ICAM-1 associated interaction with lymphocyte cells than non-inf DC, which was reported to activate T cells and B cells. They also showed higher HLA gene interaction in COPD and IPF, which is the functional gene of presenting antigen, but not observed in LUAD. Besides, we also observed that inf-DC in COPD and IPF showed a higher interaction of CCL22 and CCL3, but not in LUAD. SPP1 was reported as cancer-promoting gene in pulmonary, and the interaction associated with SPP1, such as SPP1_CD44 and SPP1_PTGER4, were detected on non-inf DC of COPD, and cDC2 of IPF, but not found in LUAD. Although the inf-DC behaved differently in chronic disease and lung cancer, we found that they shared the same pathogenic gene SEMA7A, which was reported to take part in various lung diseases,36, 37 and that was not found in normal tissue (Figure S5).
In summary, the results indicated that although inf-DC were generally more active, they behaved differently in COPD, IPF and LUAD, and the roles of inf-cDC1, inf-cDC2 and inf-mature-DC are distinct. And the performance of inf-DC in chronic lung diseases such as COPD and IPF might indicate the development of lung cancer.
3 DISCUSSION
With the development and extensive application of single-cell sequencing technologies, it is convenient to study the evolving patterns of tumour and the heterogeneity of cell population.38, 39 In this article, we carried out a comprehensive scRNA-seq analysis covering para-tumour, tumour tissues, tissues from patients with diverse chronic lung diseases and normal tissue.
We found that the DC from tissues of patients with different lung diseases clustered regularly on the UMAP, and identified inf-DC subsets from each of the three DC subtype. In previous scRNA-seq research of DC subtypes, inf-DC was not identified in cDC1 or mature-DC subsets. In this article, we confirmed an inf-DC pattern in cDC1 and mature-DC and their higher expression of pro-inflammatory cytokines and so on. We also investigated the inf-DCs function in various lung diseases. The results indicated that the role of inf-cDC1, inf-cDC2 and inf-mature-DC were totally different (Figure 4), and inf-cDC2 seemed to promote immune resistance, while the other two seemed associated with cancer development. Besides, we also observed their distinct interaction with lymphocyte cells in different lung diseases. While considering the gene SEMA7A and its role in chronic lung disease, we observed its activation in all of the lung diseases, IPF, SSC, LUAD, etc., except for normal tissue. The inf-DC in chronic diseases and lung cancer shared some similar signatures and behaviours.
More importantly, we observed a cancer associated inf-DC abundance pattern in lung cancer tissue. In fact, the inf-DC abundance pattern was highly patient dependent, some of the patients with chronic lung diseases and all of the normal tissue from LUAD patients were observed to share the same inf-DC pattern with that of lung cancer. And the percentages of such patients were very close to that reported.16, 40 In this study, we discovered that inf-DCs is present in all three of the DC subtypes. Therefore, we suspect the heterogeneity of DC pattern is a potential index to predict the risk of developing into lung cancer. However, we have not proved its reliability in real cases, and we seek to verify the inf-DC pattern in more situations and its pathological mechanism with lung cancer in future research.
4 METHODS
The datasets were collected from GSE136831, GSE128169, GSE128033, GSE136831, GSE131907_Lung_Cancer, E_MTAB_6653 and E_MTAB_6149. For better data integration, the cells with less than 200 genes expressed and genes expressed in less than three cells were excluded in each dataset. Then, each data were applied log-transformed normalisation with function ‘NormaliseData’.
We utilised the ‘FindIntegrationAnchors’ of Seurat R package (V4) to get anchors between datasets, the ‘anchor.features’ were set as 4000. Then, the datasets were integrated by ‘IntegrateData’ with default ‘CCA’ method. The integrated data object were used for downstream analyses. The integrated data were then scaled and applied principal component analysis with top 30 pcs computed. After which, the ‘FindNeighbours’ function of Seurat was employed to construct the shared nearest neighbour graph, then unsupervised cluster was performed by the ‘Find Clusters’ function. The UMAP was employed for further visualisation with default settings.
After first ground of integration, we obtained all pulmonary cell data, with 16 490 genes and 613 952 cells. Cells are divided into five main cell types, endothelial, epithelial, immune-lymphocyte (lymphocyte), immune-myeloid (myeloid) and stromal (Figure S6a).
The myeloid clusters were extracted and applied with the second ground of integration, the DC, endothelial and monocyte/macrophage clusters were identified with marker genes expression. CD1A, CLEC9A and LAMP3 for DC, VWF for endothelial, EPCAM for epithelial and CD14 and CD163 for macrophage (Figure S6b).
After all, DC clusters were extracted for the third ground integration. And we classified DC subtypes cDC1, cDC2 and mature-DC with canonical markers (XCR1, CLEC9A and CADM1 for cDC1; FCER1A, CLEC10A, CD1C, SIRPA and ITGAM for cDC2; LAMP3, CD80, CD86, CD40, CD83 and RELB for mature-DC) (Figure S6c).
AUTHOR CONTRIBUTIONS
All authors contributed to the idea, writing and editing.
ACKNOWLEDGEMENTS
The graphical abstract was drawn by Figdraw. This work was supported in part by the National Natural Science Foundation of China (82170045 to JH); Special Fund for Scientific Research of Shanghai Landscaping & City Appearance Administrative Bureau (G222410 to JH and XZ); the Translational Medicine Cross Research Fund of Shanghai Jiao Tong University (ZH2018QNB29 to JH); the Natural Science Foundation of Shanghai Municipality (16ZR1417900 to XZ); the Shanghai Pujiang Program (16PJ1405200 to XZ and 16PJ1405100 to JH); and the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZLCX20212301 to JH).
CONFLICT OF INTEREST STATEMENT
The authors declare they have no conflicts of interest.
ETHICS STATEMENT
Since the sequenced data generated from GEO were publicly available, additional ethnics committee approval was not necessary.
Open Research
DATA AVAILABILITY STATEMENT
Data will be made available on request.




