The roles of ubiquitination-mediated intrinsic apoptotic signalling in cancer therapy
Abstract
Ubiquitination, a highly versatile process of protein homeostasis, participates in the degradation of the vast majority of cellular proteins. In addition to proteolytic function, ubiquitination can also conduce to modulate cellular processes independent of proteolysis, such as signal transduction, protein trafficking, DNA repair and so on. Apoptosis modulates cell quantity via orderly removing damaged cells effectively and plays a fundamental role in the function of multicellular organisms as well homeostasis, while abnormal regulation of apoptosis can lead to various diseases, especially cancer. Since various tumour cells intrinsically elude apoptotic elimination, emerging strategies targeting induction of apoptosis have become great potential and often necessary cancer therapeutic approaches. Numerous researches have shown that ubiquitination can facilitate or inhibit apoptosis by utilising its proteolytic or non-proteolytic functions aiming at controlling the levels of key proteins. In this review, we focused on intrinsic apoptosis and summarised how diverse types of ubiquitination regulate intrinsic apoptosis through E3 ligases and deubiquitinating enzymes. Given the mechanisms of ubiquitin-mediated regulation of intrinsic apoptosis and their physiological relevance, we hope that it will supply more therapeutic strategies for cancer.
Abbreviations
-
- FDA
-
- Food and Drug Administration
-
- NF-κB
-
- Nuclear factor kappa-BMM:Multiple myeloma
-
- SS
-
- Sjogrensyndrome
-
- SLE
-
- Systemic Lupus Erythematosusd
-
- ATP
-
- Deoxyadenosine triphosphate
-
- T-ALL
-
- T-cell acute lymphocytic leukemia
-
- ARTS
-
- Apoptosis related protein in TGF-beta signaling pathway
-
- CSC
-
- Cancer stem cells
-
- HPV
-
- Human papilloma virus
-
- GPER /MAPK/ERK
-
- G-protein estrogen receptor/ mitogen-activated protein kinase/ extracellular signal-regulated kinase
-
- SOCS
-
- The suppressors of cytokine signalling
-
- TAM
-
- tamoxifen
-
- JNK
-
- The c-Jun NH(2)-terminal kinases
-
- TRAIL
-
- the tumour necrosis factor-related apoptosis-inducing ligand
-
- AIF
-
- apoptosis-inducing factor
-
- SMAC/DIABLO
-
- The second mitochondria-derived activator of caspase/direct inhibitor of apoptosis protein-binding protein with low Pi
-
- HTRA2/OMI
-
- High-temperature-requirement A2
-
- ENDOG
-
- Endonuclease
-
- GAGS
-
- human gastric adenocarcinoma cell line
-
- CACI
-
- Cdk-Associated Cullin1
-
- BAX
-
- BCL-2-Associated X Protein
-
- TRIM9
-
- Tripartite motif-containing 9
-
- XIAP
-
- X-linked inhibitor of apoptosis
-
- PARK2
-
- Biallelic Parkin
-
- FBXO4
-
- F-Box protein 4
-
- MCL1
-
- Myeloid Cell Leukemia 1
-
- MARCH5
-
- Mitochondrial ubiquitin ligase membrane-associated
-
- RING-CHNOXA
-
- the proapoptotic BH3-only protein
-
- VDAC2
-
- Voltage-Dependent Anion Channel Protein 2
-
- SCF
-
- The SKP1, CUL1, F-box protein
1 INTRODUCTION
Protein homeostasis, a dynamic equilibrium process in which the body regulates the balance of intracellular proteins synthesis, folding, trafficking, assembly and degradation through various control pathways, is a pivotal task for correct cellular function. Actually, cells have developed plenty of strategies to achieve it under stress, especially the ubiquitin–proteasome system (UPS) in which the degradation of the vast majority of cellular proteins are involved.1 UPS has two main functions: One is to advocate cell quality by degrading damaged and misfolded proteins; the other is to possess the basic life activities of the cell by undermining proteins with specific functions. These two different effects ultimately ensure the proper functioning of tissues and organs like DNA repair, stress responses and cell proliferation for apoptosis.
Apoptosis is a vital physiological process in multi-cellular organisms, elicited by varieties of cellular signalling processes to clean up any unwanted or damaged cells and thus to maintain cellular and tissue homeostasis.2 Aside from the pivotal role in normal physiology, numerous researches on apoptosis have demonstrated that aberrations of apoptosis can be associated with the pathogenesis of various human diseases.3 Take cancer for instance, in the tumour microenvironment, cells lose their ability to receive apoptosis signals, leading to the turbulence of homoeostatic balance between newly generated cells and cell death. It was established that a normal cell transforming into a malignant one with subsequently uncontrolled proliferation is a hallmark of cancer. Tumour cells are often found to over-express anti-apoptosis proteins, which inhibit apoptotic cascade and finally promote cancer progression. For example, B-cell lymphoma 2 (Bcl-2) overexpression leads to inhibition of TRAIL-induced apoptosis in neuroblastoma, glioblastoma and breast carcinoma cells,4 while overexpression of tripartite motif 14 (TRIM14) promotes breast cancer cell proliferation by inhibiting apoptosis.5 Thus, suppression of apoptosis is the culprit that malignant cells start continuous proliferation and ultimately lead to diseases deterioration even cancer. Obviously, dwindled apoptosis promotes tumour development, which made targeting the cell apoptosis machinery a tractable and promising cancer therapeutic strategy. Therefore, therapeutic strategies targeting molecules involved in apoptotic resistance represent a valid approach to be pursued in order to restore cancer cells' sensitivity to apoptosis and overcome the ineffectiveness of the treatments.
Recent studies have shown that a large number of E3 ubiquitin (Ub) ligases and deubiquitinating enzymes (DUBs) are involved in a complex network of interactions, regulating plenty of key molecules of the apoptotic pathway. In this review, we will focus on intrinsic apoptosis, discussing and summarising studies of the recent decades about key Ub-specific enzymes and their clinical application in cancer therapy.
2 Ub, A HIGHLY VERSATILE REGULATOR OF PROTEIN HOMEOSTASIS
2.1 Proteolytic and non-proteolytic functions of ubiquitination
Ubiquitination is a ubiquitously dynamic post-translational modification (PTM) involved in regulating a myriad of important cellular functions including cell cycle, DNA repair and membrane trafficking in almost all eukaryotes.6 With the stable and conserved structure, Ub, the 76-amino-acid protein, acts by conjugating lysine (Lys) residues to a variety of cellular proteins, including misfolded or abnormal proteins, as well as proteins that require stock regulation and instability.7 Addition of Ub to target protein is carried out by multi-enzyme complex, which is catalysed successively by three specific enzymes, Ub-activating enzymes (E1), Ub-conjugating enzymes (E2) and Ub-ligases (E3).8 Free Ub is activated by forming a thioester bond with the E1 Ub-activating enzyme in an adenosine triphosphate (ATP)-dependent manner. Subsequently, Ub is transferred to an E2 Ub-conjugating enzyme and ultimately combines with target proteins through the actions of an E3 Ub-ligase enzyme (Figure 1A).8, 9 In the UPS, the substrate with Ub tag prefers to be recognised by the proteasome cap and ultimately transferred to the core lumen of the proteasome for degradation.
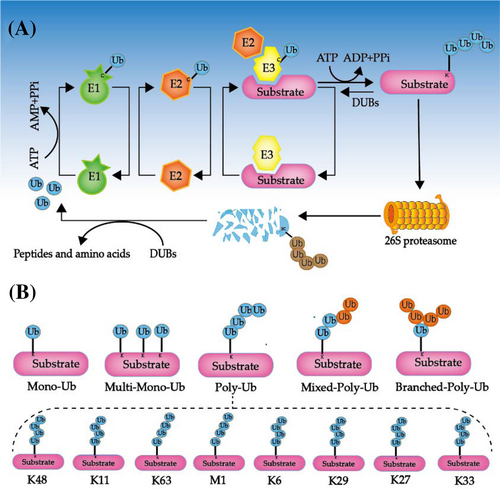
The degrading function of ubiquitination acts as a fundamental role in protein homeostasis, but it does not stop there. It is now well-established that the ubiquitination effects do not solely target protein for degradation but can also conduce to modulate cellular processes independent of proteolysis, such as signal transduction, protein trafficking, DNA repair, vastly hinging on the types of Ub modifications.10 Diverse types of ubiquitination modifications have been found in recent years (Figure 1B). The simplest type is monoubiquitylation (mono-Ub) by which a single Ub molecule is attached, while a single Ub molecule can alternatively tag with several Lys residues, facilitating multiple mono-Ub, namely, multiubiquitylation (multi-Ub).11 Because the Ub moiety contains seven Lys (K) residues (K6, K11, K27, K29, K33, K48, K63) and methionine at the amino-terminus (M1) itself, Ub molecules can form different types of chains in an iterative process, referred to as polyubiquitylation (poly-Ub).12 All of these types play key roles in apoptosis signalling via distinct modulation strategies. It is obvious that Lys11 and Lys48-linked poly-Ub chains represent a signal for proteasomal degradation,8, 13 whereas Lys63-linked chains are more typically related to the resistance of DNA damage, endocytosis and signal transduction, which M1-Liner sometimes can coordinate with in NF-κB.14, 15 All of these types play key roles in apoptosis signalling via distinct modulation strategies.
2.2 E3 Ub ligase, a key modulator of ubiquitination
Indeed, E1--E2--E3 cascade reaction plays a pivotal role in the ubiquitination process, among which E3 determines the specific recognition of substrate proteins. While the E1 family contains two members, and E2 families have around 40 members, the E3 Ub ligase family is constituted of approximately 700 members. These E3 enzymes are generally grouped into three major classes: (a) the ‘really interesting new gene’ (RING) domain-containing E3s, which act as scaffolds for E2 to directly transfer Ub to target proteins without forming thioester bonds16; (b) the 'homologous to E6-AP carboxyl terminus' (HECT) domain-containing E3s, which form a catalytic cysteine (Cys)-dependent intermediate with Ub before conjugation to the target substrate17; and (c) the 'RING-between-RING' E3s, which function as hybrids of RING and HECT E3s, catalyzing Ub conjugation by directly forming a thioester intermediate with a Cys of RING2 domain.18
2.3 Leader of reverse ubiquitination: DUBs
Like other PTMs, ubiquitination is reversible and can be opposed by a large group of proteases called DUBs.19 DUBs are a large protease family, in which the mammalian genome encodes approximately 100 different DUBs. In humans, DUBs can be classified into eight subfamilies based on their sequence and structure similarity: Ub-specific proteases (USPs), ovarian tumour proteases, Ub C-terminal hydrolases (UCHs), Machado–Joseph disease protein domain proteases, JAP1/MPN/Mov34 metallopeptidases, the motif interacting with Ub-containing novel DUB family, the monocyte chemotactic protein-induced protein and zinc-finger and UFM1-specific peptidases.20-22 DUBs facilitate protein homeostasis and sustain Ub levels through multiple strategies including fabrication of Ub precursors or adducts to participate in signal transduction, hydrolyzation of Ub from target substrate into the circulation reuse and regulation of cellular degradation pathways.23, 24
3 REGULATION OF INTRINSIC APOPTOSIS BY UBIQUITINATION
In 1972, Kerr, Wyllie and Currie first used the term 'apoptosis' to describe a type of cell death, which appears to play a complementary but opposite role to mitosis in the regulation of animal cell populations. Simultaneously, they described the involvement between cellular apoptosis and the elimination of potentially malignant cells, hyperplasia and cancer progression.2 Subsequent studies have uncovered the mysterious veil of apoptosis, a genetically controlled process of programmed death, in which a cell quits proliferation and differentiation and instead spontaneously enters an orderly process that ultimately results in the controlled death of the cell without releasing its contents into the ambient environment.25
Revealing the detailed procedure and mechanisms of apoptosis is of great significance for us to understand the pathogenesis of conditions initiated by disordered apoptosis, thereby may in turn help in the research and development of new drugs or treatment strategies that target certain apoptotic genes or pathways in diseases. The core component of the apoptosis pathway is caspase, which can be activated by three distinct apoptosis signalling: intrinsic mitochondrial pathway, endoplasmic reticulum pathway and the extrinsic pathway, which is also known as the death receptor pathway.26 Each pathway interacts with another and regulates the process of apoptosis together. In recent years, increasing studies have found that ubiquitylation plays an important role in the upstream or downstream of apoptosis, and the high selectivity of E3 enzymes and DUBs to substrates is an important reason for its specificity. How UPS influence the extrinsic apoptosis pathway has been reviewed elsewhere.27 Thus, we focused on the latest developments in the study of E3 ligase and DUBs, which contribute to intrinsic apoptosis pathways.
Just as its name implies, the mitochondria-dependent pathway is instigated by intracellular signals that converge at the mitochondrial level in response to different apoptotic stimulating conditions, such as irreparable genetic damage, cell hypoxia and severe oxidative stress.28 Regardless of the stimuli, this pathway is the result of changed mitochondrial outer membrane permeability (MOMP) and the release of pro-apoptotic molecules such as cytochrome-c (Cyt-C), AIF, SMAC/DIABLO, HTRA2/OMI, ENDOG into the cytoplasm, which can be closely regulated by the apoptosis-related factors like Bcl-2 family proteins.26, 29 Released Cyt-C activates caspase-9 via the formation of an apoptosome complex, which is composed of Cyt-C, Apaf-1 and pro-caspase-9. Subsequently, activated caspase-9 further activates caspase-3 and caspase-7, ultimately leading to apoptosis29 (Figure 2).
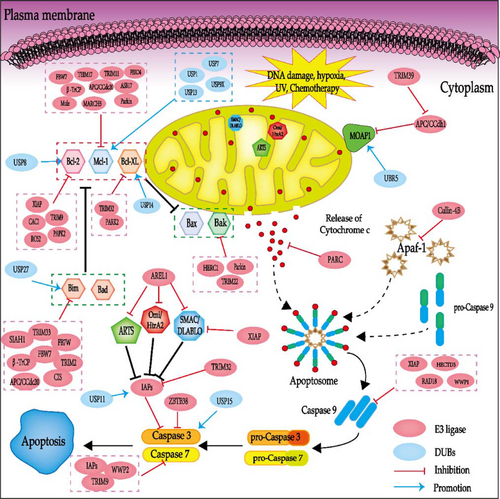
3.1 The Bcl-2 family proteins
The Bcl-2 family of proteins act as pivotal modulators of the mitochondrial pathway of cellular apoptosis, comprising both pro-apoptotic proteins like Bak, Bax, Bid, Bim, Bad, Bik, Bmf, Noxa, Puma and anti-apoptotic proteins including Bcl-2, Bcl-xL, Bcl-W, A1 and Mcl-1, playing key roles in cancer cell survival.30, 31 While the anti-apoptotic proteins are mainly located in the inner membrane of cells, the majority of the pro-apoptotic proteins are vastly distributed at the outer membrane of mitochondria in the cytoplasm. The pro-apoptotic proteins facilitate apoptosis by initiating the release of Cyt-C, whereas the anti-apoptotic proteins function by inhibiting such release. However, it is the balance between the pro-apoptotic and anti-apoptotic proteins rather than the absolute quantity that dictates the apoptosis whether it would be initiated or not.32 A large number of studies have shown that ubiquitination plays a significant role in the regulation of the balance, disturbance or correction (Table 1).
| Substrate | E3/deubiquitinating enzymes (DUBs) | Function | Effect on apoptosis | Associated diseases | Ref. | |
|---|---|---|---|---|---|---|
| B-cell lymphoma 2 (Bcl-2) family proteins | ||||||
| Bcl-2 | E3 | TRIM9 | TRIM9 knockdown elicited the reduction of Bcl-2 expression | Inhibit | Lung cancer | 35 |
| E3 | CAC1 | CAC1 might protect AGS cells from apoptosis by altering BCL2/BAX ratio. | Inhibit | Gastric carcinoma | 33 | |
| E3 | XIAP | Ubiquitinates and degrades Bcl-2 at lysine (Lys) 17 | Promote | Haematological malignancies, solid tumours | 36 | |
| E3 | PARK2 | Degrades BCL-2 | Promote | Breast cancer | 38 | |
| E3 | Ro52 | Represses Bcl-2 production | Promote | SS, SLE | 37 | |
| DUBs | USP8 | Regulates the Bcl-2/Bax axis and caspase cascade | Inhibit | Bile duct cancer | 34 | |
| Mcl-1 | E3 | FBW7 | Reducing Mcl-1 expression | Promote | T-ALL | 98 |
| E3 | Mule | Reducing Mcl-1 expression | Promote | Unclear | 99 | |
| E3 | β-TrCP | Reducing Mcl-1 expression | Promote | Lung cancer | 100 | |
| E3 | TRIM17 | Reducing Mcl-1 expression | Promote | Unclear | 101 | |
| E3 | APC/CCdc20 | Controls Mcl-1 instability | Promote | Unclear | 102 | |
| E3 | FBXO4 | Promoting Mcl-1 ubiquitination and degradation | Promote | Lung cancer | 40 | |
| E3 | TRIM11 | Reduces the stability of MCL1 | Promote | Lung cancer | 42 | |
| E3 | MARCH5 | Driving degradation | Promote | Prostate cancer | 43 | |
| E3 | ASB17 | Degrading MCL1 | Promote | Unclear | 44 | |
| E3 | Parkin | Degrading MCL1 | Promote | Parkinson's disease | 45 | |
| DUBs | USP7 | Stabilises MCL1 | Inhibit | Lung cancer | 49 | |
| DUBs | USP9X | Removes degradative Lys48-linked poly-ubiquitin chains from Mcl-1 | Inhibit | Follicular lymphoma | 46 | |
| DUBs | USP1 | Stabilises the expression of anti-apoptotic proteins Bcl-2 and Mcl-1 | Inhibit | Colorectal cancer | 50 | |
| DUBs | USP13 | Stabilises MCL1 and modulate tumour cell sensitivity to BH3 mimetic inhibitors | Inhibit | Cervical cancer | 47, 48 | |
| Bcl-XL | E3 | PARK2 | Binds to and ubiquitinates BCL-XL protein to control apoptosis | Promote | Unclear | 74 |
| E3 | TRIM32 | Overexpression increased the mRNA level | Inhibit | Glioma | 103 | |
| DUBs | USP14 | Interacts with Bcl-xl and upregulates its protein level | Inhibit | Multiple myeloma | 104 | |
| Bim | E3 | Fbw7 | Increasing Bim expression | Inhibit | B cell cancer | 54 |
| E3 | Cullin2/ElonginB-CIS | Degrade BimEL | Inhibit | Ovarian, colon and breast cancer | 61, 62 | |
| E3 | TRIM2 | Mediates the MAPK-dependent ubiquitination of Bim | Inhibit | Breast cancer | 53, 60 | |
| E3 | SIAH1 | Regulates the level of Bim by JNK pathway | Promote | Breast cancer | 55 | |
| E3 | βTrCP1 | Mediates stability of BimEL | Inhibit | Unclear | 56 | |
| E3 | APC/CCdc20 | Targets Bim for ubiquitination and destruction | Inhibit | T cell leukaemia | 57 | |
| E3 | TRIM33 | Blocks enhancer-mediated Bim activation | Inhibit | B lymphoblastic leukaemia | 58 | |
| DUBs | USP27x | Stabilise Bim | Promote | Unclear | 63 | |
| Bak | E3 | HERC1 | Ubiquitinates at residue K113 and degrades BAK via a BH3 domain | Inhibit | Non-melanoma skin cancer | 66 |
| E3 | Parkin | Ubiquitinates BAK at residue K113 to inhibit BAK oligomerisation | Inhibit | Parkinson's disease | 67 | |
| E3 | TRIM22 | Regulates Bak oligomerisation | Inhibit | Sepsis | 105 | |
| cytochrome c (Cyt-C) pathway | ||||||
| MOAP-1 | E3 | APC/CCdh1 | Ubiquitylates and degrades MOAP-1 during G1 | Inhibit | Unclear | 70 |
| E3 | UBR5 | Elicits ubiquitylation of MOAP-1 and inhibits MOAP-1 stability | Inhibit | Ovarian cancer | 72 | |
| APC/CCdh1 | E3 | TRIM39 | Ubiquitylates and degrades APC/CCdh1 | Promote | Unclear | 71 |
| Cyt-C | E3 | PARC | Mediates ubiquitination and degradation of Cyt-C | Inhibit | Neuroblastoma, glioma | 76 |
| Smac | E3 | XIAP | Degrade Smac | Inhibit | Unclear | 77 |
| Apaf-1 | E3 | Cullin-4B | Modulates Apaf-1 ubiquitination at Lys 224 to control caspase-9 activity | Inhibit | Unclear | 84 |
| Caspase-9 | E3 | HECTD3 | Binds and ubiquitinates caspase-9 at Thr-157 | Inhibit | ESCC | 79 |
| E3 | XIAP | Inhibits the homodimerisation of caspase-9 by binding to its BIR3 domain | Inhibit | Unclear | 85, 106 | |
| E3 | RAD18 | Activates caspase-9 and caspase-3 | Promote | Rectal cancer | 82 | |
| E3 | WWP1 | WWP1 knockdown is related to caspase-9 activation, cleavage of PARP | Inhibit | Breast cancer | 83 | |
| Caspase-3 | E3 | cIAP1 | Ubiquitinates and degrades caspase-3 and caspase-7 | Inhibit | Unclear | 87 |
| E3 | ZBTB38 | Participates in the negative regulation of apoptosis | Inhibit | Unclear | 95 | |
| E3 | XIAP | Inhibits caspase-3 and caspase-7 by binding the N terminus of the BIR2 domain | Inhibit | Unclear | 86 | |
| E3 | TRIM9 | Reduces expression of cleaved-caspase-3 | Inhibit | Uterine leiomyoma | 107 | |
| DUBs | USP15 | Activates caspase-3 | Promote | Ovarian cancer | 96 | |
| Caspase-7 | E3 | cIAP1 | Ubiquitinates and degrades caspase-3 and caspase-7 | Inhibit | Unclear | 37 |
| E3 | WWP2 | Reduces expression of caspase-7, increases expression of Bcl-2 | Inhibit | Liver cancer | 97 | |
| E3 | XIAP | Inhibits caspase-3 and caspase-7 by binding to the N terminus of the BIR2 domain | Inhibit | Unclear | 86 | |
| XIAP | E3 | TRIM32 | Ubiquitinates XIAP in a ‘really interesting new gene’ finger-dependent manner | Promote | Muscular dystrophy, epithelial carcinogenesis | 92 |
| E3 | USP11 | Binds to XIAP through Leu207 on XIAP | Promote | Breast cancer | 93 | |
| Smac, Omi, ARTS | E3 | AREL1 | Interacts with and ubiquitinates SMAC, HtrA2 and ARTS | Inhibit | Unclear | 94 |
- Abbreviations: cIAP1, cellular inhibitor of apoptosis proteins; CIS, cytokine-inducible Src homology 2 domain-containing protein; ESCC, oesophagal squamous cell carcinoma; HECT, homologous to E6-AP carboxyl terminus; MOAP, modulator of apoptosis; TRIM, tripartite motif; USP, ubiquitin-specific protease; MM, Multiple myeloma; SS, Sjogrensyndrome; SLE, Systemic Lupus Erythematosus; dATP, Deoxyadenosine triphosphate; T-ALL, T-cell acute lymphocytic leukemia; ARTS, Apoptosis related protein in TGF-beta signaling pathway; CSC, Cancer stem cells; HPV, Human papilloma virus; GPER/MAPK/ERK, G-protein estrogen receptor/ mitogen-activated protein kinase/extracellular signal-regulated kinase; SOCS, The suppressors of cytokine signalling; TAM, tamoxifen; JNK, The c-Jun NH(2)-terminal kinases; TRAIL, the tumour necrosis factor-related apoptosis-inducing ligand; AIF, apoptosis-inducing factor; SMAC/DIABLO, The second mitochondria-derived activator of caspase/direct inhibitor of apoptosis protein-binding protein with low Pi; HTRA2/OMI, High-temperature-requirement A2; ENDOG, Endonuclease G; AGS, human gastric adenocarcinoma cell line; CACI, Cdk-Associated Cullin1; BAX, BCL-2-Associated X Protein; XIAP, X-linked inhibitor of apoptosis; PARK2, Biallelic Parkin; FBXO4, F-Box protein 4; MCL1, Myeloid Cell Leukemia 1; MARCH5, Mitochondrial ubiquitin ligase membrane-associated RING-CH; NOXA, the proapoptotic BH3-only protein; VDAC2, Voltage-Dependent Anion Channel Protein 2; SCF, The SKP1, CUL1, F-box protein.
3.1.1 Bcl-2
Anti-apoptotic protein, like Bcl-2, has been reported over-expressed in different haematological malignancies and solid tumours. A large number of studies have shown that the interaction between E3 ligase/DUBs and Bcl-2 plays an important role in promoting or inhibiting the apoptosis of tumour cells. It is obvious that the Bcl-2/BAX ratio governs the sensitivity of cells to apoptotic stimuli. In the process of cisplatin-induced apoptosis, CAC1 might be conducive to the occurrence of AGS cell apoptosis by altering Bcl-2/BAX ratio because CAC1 silencing brought out a prominent increase of BAX and a decrease of Bcl-2.33 In addition, recent research showed upregulation of USP8 can also decrease apoptosis in Hucct-1 cells, while silencing of USP8 promote apoptosis by regulating the Bcl-2/Bax axis and caspase cascade as well as inhibiting activation of the Akt signalling pathway by decreasing the phosphorylation level of Akt and upregulating p53 expression.34 Furthermore, Wang et al. proved that TRIM9 suppressed apoptosis. In lung cancer cells, TRIM9 knockdown elicited the reduction of Bcl-2 expression and concomitantly activation of caspase-7 and caspase-9.35 In addition to the inhibiting effect, Ub-specific enzymes can also promote apoptosis. XIAP functions as an E3-ligase for ubiquitylating Bcl-2 at Lys 17. Interestingly, the generally accepted view is that XIAP has an anti-apoptotic function, but Natalia et al. indicated that ARTS acts as a distinct Bcl-2 antagonist that brings Bcl-2 into a ternary complex with XIAP, thereby stimulating UPS-mediated degradation of Bcl-2 to promote apoptosis.36 E3 ligase Ro52 function is connected to various environmental stimuli and is important in inducing apoptosis by repressing Bcl-2 protein production.37 Meanwhile, ubiquitination may play an important role in enhancing sensitivity to cancer drugs. Chen et al. found that PARK2 degraded phospho-Bcl-2 in an E3 ligase-dependent manner, activating apoptotic mitochondrial pathways and ultimately mediating anti-microtubule drug sensitivity.38
3.1.2 Mcl-1
As an anti-apoptotic member of the Bcl-2 family, the mechanism underlying the effect of Mcl-1 in apoptosis has been extensively studied, among which its ubiquitination was identified to be regulated by different enzymes. Barbara et al. summarised the influence of Mule, SCFβ-TrCP, SCFFbw7, APC/CCdc20, Trim17 on Mcl-1 in their review,39 and we will not repeat it too much here. It has been reported that Mcl-1 is frequently over-expressed in lung cancer. Feng et al. found that FBXO4 inhibits lung cancer cell survival by promoting Mcl-1 ubiquitination and degradation in lung cancer cell lines and lung cancer patient samples.40 Studies have shown that TRIM11 may have an important role in promoting apoptosis.41 Recently, Li et al. speculated that TRIM11, a novel discovered Ub E3 ligase of MCL1, may induce apoptosis by reducing the stability of MCL1 and leading to the decrease of its protein levels, while SUMOylation of MCL1 inhibits TRIM11-mediated MCL1 ubiquitination.42 In prostate cancer cells, mitochondria-associated Ub ligase MARCH5 serves as the mediator of NOXA-dependent MCL1 degradation.43 ASB17 positively regulates cell apoptosis by promoting the ubiquitylation and degradation of BCLW and MCL1.44 Mitochondrial depolarisation promotes Parkin- and PTEN-induced kinase 1 (PINK1)-dependent polyubiquitination of multiple proteins on mitochondrial outer membranes, resulting in the removal of defective mitochondria via mitophagy. Upon mitochondrial depolarisation, the Bcl-2 family member Mcl-1 underwent rapid Parkin- and PINK1- dependent polyubiquitination and degradation, which sensitised towards apoptosis via the opening of the Bax/Bak channel.45
Besides, ubiquitination is a reversible event that can be counteracted by specific enzymes. In follicular lymphomas, USP9X has been identified to remove degradative Lys48-linked poly-Ub chains from Mcl-1, thereby stabilising it and leading to apoptosis resistance and promoting tumour cell survival.46 However, Zhang et al. provided evidence that USP9X knockdown did not remarkably alter MCL1 protein levels in multiple lung and ovarian cancer cell lines. Another DUBs, USP13, was found to be a bona fide DUB for MCL1, stabilising MCL1 and modulating tumour cell sensitivity to BH3 mimetic inhibitors.47 Subsequently, Morgan et al. confirmed that USP13 is highly expressed in cervical cancer cell lines and cervical cancer tissue, functionally deubiquitinating and stabilising the pro-survival protein of Mcl-1.48 Furthermore, Yang et al. speculated that USP7 stabilises MCL-1 protein during the tumourigenic process by the evidence that USP7-mediated MCL-1 upregulation enhances arsenic and BaP co-exposure-induced CSC-like property and tumourigenesis.49 In addition, knockdown of USP1 induced growth arrest at G2/M of cell cycle and reduced the expression of anti-apoptotic proteins Bcl-2 and Mcl-1 in cultured CRC cells.50
3.1.3 Bim
Bim, a BH3 domain-containing protein, acts as a pro-apoptosis modulator during apoptosis progression. Decline in Bim expression was found to be related to tumour development, while its overexpression was shown to have a tumour suppressive function that restrained tumour growth and drug resistance in renal carcinoma and non-small cell lung carcinoma.51, 52 Recently, eight E3 ligases, TRIM2,53 Fbw7,54 SIAH1,55 β-TrCP1,56 APC/CCdc2,57 TRIM33,58 Cullin2/Elongin B-cytokine-inducible Src homology 2 domain-containing protein (CIS)59 were identified to contribute to the ubiquitination and degradation of Bim in different tumour cells.
The TRIM proteins, one of the subfamilies of the RING-type E3 Ub ligases, are involved in the development of human cancers. Wang et al. stimulated that TRIM33 suppresses apoptosis in murine B-ALL cells by blocking enhancer-mediated Bim activation.58 Using a proteomics approach, Thompson et al. implicated that TRIM2 may mediate the p42/p44 MAPK-dependent monoubiquitination of Bim in rapid ischemic tolerance.53 Subsequently, Yin et al. provided evidence that activated GPER/MAPK/ERK triggered enhanced TRIM2 protein levels and affected the binding between TRIM2 and Bim, which resulted in a reduced Bim in TAM-resistant breast cancer cells, but the Ub-binding site are still unclear.60 In addition to TRIM families, some E3s have been reported to inhibit apoptosis. CIS and a SOCS box domain-containing E3 ligases, forms an E3 multi-subunit complex with Elongin B/C and Cullin2. In both ovarian and breast cancer cell lines and specimens, Zhang et al. found the E3 multi-subunit complex can degrade BimEL protein and determine BimEL levels by means of its bridging with BimEL, entailing its potential role in the development of drug resistance in tumours.61 Grazia obtained similar result in HCT116 human colon cancer cells.62 APCCdc20 was reported to interact with BimEL and govern its proteasomal degradation and rescue from Ub-dependent degradation, while inhibition of it conduced to apoptosis induction and chemo-radiation sensitisation in cancers of different origins.57 Besides, some other Ub-specific enzyme was found to promote apoptosis. Fbw7 can increase Bim expression in autoimmune diseases,54 while βTrCP1 and USP27x initiate apoptosis by controlling the stability of Bim.56, 63 In human breast cancer tissues and cells, overexpression of SIAH1 induce apoptosis of breast cancer cells by upregulating the expression of Bim through the activation of the JNK pathway.55 In contrast, the reduction of SIAH1 in these cells increased p-ERK expression and vice versa. Therefore, SIAH1 may inhibit the invasion of cancer cells through the ERK pathway.55
3.1.4 Bax/Bak, Bcl-XL
The intrinsic mitochondrial apoptotic pathway is required for efficient chemotherapeutic killing of cancer cells and is initiated through a nodal point in the decision of a cell to undergo apoptosis, which is the BH3-only protein activation of Bax/Bak-mediated MOMP. The BAK function relies on the hydrophobic surface groove on BAK to a great extent, as this region not only permits its interaction with BH3-only proteins but also is essential for BAK homo-dimerisation.64, 65 Holloway et al. observed that HERC1 contains a putative BH3 domain that can bind to the exposed groove of BAK.66 In HPV-infected cells, HERC1 facilitates the degradation of BAK, which is dephosphorylated at Y108 and undergoes the N-terminal conformational change by binding and ubiquitinating BAK at residue K113 via a BH3 domain, thus inhibiting mitochondrial apoptosis to promote cell survival.66 Interestingly, in addition to promoting BAK degradation, another E3 ligase, Parkin, mediates non-degradative ubiquitination of BAK through obscuring the hydrophobic surface groove to inhibit BAK function.67 Bernardini et al. proposed Parkin that not only can ubiquitinate BAK at residue K113 to inhibit BAK oligomerisation but can also ubiquitinate VDAC2 to limit BAX association with mitochondria, thereby inhibiting apoptosis.67
The modulator of apoptosis (MOAP-1), which contains a BH3-like motif, has been proved to be an effector for Bax function to promote Bax mitochondrial translocation and activation.68, 69 Initially, Fu et al. found that inhibition of Ub-mediated degradation of MOAP-1 promotes Bax function in mitochondria, in spite of the E3 Ub ligase responsible for MOAP-1 ubiquitination is unclear.70 Subsequently, Huang et al. determined APC/CCdh1 can ubiquitylate and degrade MOAP-1 during G1, and this effect can be inhibited by Trim39 acting on the APC/C, but the operating mechanism was less understood.71 Recently, they reported UBR5, a novel E3 Ub ligase for MOAP-1, elicits ubiquitylation of MOAP-1 and inhibits MOAP-1 stability dependent on the cell cycle (from late S phase to G2) by cooperating with its interactor Dyrk2 kinase.72 In ovarian cancer, UBR5, Dyrk2 and APC/CCdh1 were all upregulated prior to cisplatin treatment, while MOAP-1 was downregulated. UBR5 depletion sensitised cultured ovarian cancer cells to cisplatin-induced Bax activation, and this Bax activation was dependent on MOAP-1, indicating that UBR5 as a new regulator of MOAP-1 is implicated in this regulatory network in the sensitivity of ovarian cancers to cisplatin treatment.72
It was investigated that BCL-XL suppresses the pro-apoptotic functions of BAX and BAD by binding to their BH3 domains, blocking their translocation from the cytosol to the mitochondria; nevertheless, this effect can be counteracted by the BH3-only proteins.73 Previous research showed that BCL-XL overexpresses in plenty of cancer cells such as breast cancer, lymphomas and non-small-cell lung cancer, involved in a tumour cell's ability to escape apoptosis. Through pan-cancer analysis and multiple approaches, Gong et al. found that PARK2 helps regulate apoptosis via the binding to and ubiquitinating BCL-XL protein.74 However, in glioma tumour cells, overexpressed TRIM32 increased the mRNA level of anti-apoptotic gene Bcl2, BCL2L10, Bcl-xL, Bcl-w, Mcl1 and BCL2A1, thus inhibiting apoptosis. In multiple myeloma, USP14 played a negative role in cell apoptosis by interacting with BCL-XL and upregulating its protein level, but the underlying mechanism is much less explored.
3.2 Cyt-C
The release of Cyt-C is likely to be the key event in the process of apoptosis, triggering the formation of the apoptosome complex, resulting in downstream activation of effector caspases, while other released proteins, Smac/DIABLO and Omi/HtrA2, enhance the effect by blocking inhibitor of apoptosis proteins (IAP).75 Recently, relevant studies have provided evidence that cancer cells evaded apoptosis by regulating the intracellular levels of Cyt-C. Using an unbiased siRNA screen, Gama et al. indicated that PARC/CUL9, an E3 ligase with RING and Cullin domains, targeted and degraded Cyt-C.76 Overexpression of PARC reduced the abundance of mitochondrially released cytosolic Cyt-C in various cancer cell lines, while its knockdown increased the quantity of Cyt-C and decreased viability in response to stress, suggesting that PARC-mediated ubiquitination and degradation of Cyt-C is a strategy engaged in the apoptosis resistance of cancer cells.76 Additionally, Brady et al. suggested a general mechanism to lower the mitochondrial apoptotic potential via intramitochondrial degradation of Smac.77 In response to mitochondrial depolarisation, XIAP activates Bax-mediated MOMP, permitting delayed Cyt-C release, while the mitochondria XIAP selectively degrades its inhibitor Smac through lysosome- and proteasome-associated pathway.77
3.3 Caspase
The caspases, a family of Cys proteases, are common death effector molecules that cleave different substrates in the apoptotic process. Activation of caspases can be initiated via the (intrinsic) mitochondrial pathway or the (extrinsic) death receptor pathway. When cells receive apoptotic stimuli, mitochondria releases Cyt-C, which then binds to Apaf-1, together with dATP, assembles into a caspase-activating complex named the apoptosome that recruits caspase-9 via interaction with homophilic caspase recruitment domain and in turn activates caspase-9.75 Cleaved caspase-9 further cleaves executioner caspases such as caspase-3 and caspase-7 to initiate a caspase cascade that leads to apoptosis.78 Accumulative evidence hold that the caspases family are modulated by ubiquitination, which plays a pivotal role in apoptosis resistance of cancer progression.
In the case of human oesophagal squamous cell carcinoma (ESCC), with its Thr-157 phosphorylation by ERK, HECTD3 binds and ubiquitinates caspase-9, inhibiting caspase-9 oligomerisation and association with Apaf-1, resulting in suppression of caspase-9 activation and inhibition of apoptosis.79 Moreover, in vitro assay certified that HECTD3 facilitates cell survival in ESCC and promotes tumour growth in a xenograft mouse model.79 It is obvious that the high expression of RAD18 serves to be resistant to chemotherapy or radiotherapy in numerous human cancers.80, 81 In rectal cancer cells, inhibited RAD18 upregulates cellular apoptosis by driving caspase-9/3-dependent apoptotic pathway, hence leading to the escalation of cell radiosensitivity and 5-Fu susceptibility.82 In breast epithelial cell lines, RNAi-mediated WWP1 E3 ligase knockdown is related to caspase-9 activation, cleavage of PARP and activation of apoptosis, suggesting that genomic aberrations of WWP1 may contribute to the pathogenesis of breast cancer.83 Additionally, Apaf-1 is known as a component that constitutes apoptosome with released Cyt-C, thus activating caspase-9 to facilitate apoptosis. Ohta et al. indicated that Cullin-4B E3 ligase modulates Apaf-1 ubiquitination at Lys 224 to control caspase-9 activity and inhibit apoptosis.84
IAP, with E3 ligase activity, is widely expressed in tumours and considered a potential drug target. X-linked IAP, the prototypical IAP in mammals, whose ability to bind to and suppress the initiator caspase-9 and the effector caspase-3 and caspase-7 that mediate apoptotic cell death was the primary focus of scientists after its initial discovery. XIAP binds to caspase-9 with its BIR3 domain and inhibits the homodimerisation of caspase-9,85 while XIAP combines with and inhibits caspase-3 and caspase-7 by binding to the N-terminus of the BIR2 domain.86 Besides, Choi et al. demonstrated that additional IAP family members, cellular IAP1 (cIAP1), binds to and ubiquitinates caspase-3 and caspase-7, targeting them for proteasome-dependent degradation, thereby suppressing apoptosis.87 Properly, proteasome-mediated degradation of IAPs may be an important regulatory step through which cells have received an apoptotic signal and eventually progressed to cell death. Previous research have shown five regulators of XIAP: XIAP-associated factor (XAF1),88 Smac/DIABLO,89, 90 Omi/HtrA2,91 TRIM3292 and USP11.93 As an E3 Ub ligase, TRIM32 interacts with and ubiquitinates XIAP in a RING finger-dependent manner.42 While USP11 directly binds to XIAP through Leu207 on XIAP and stabilises it by deubiquitylation of XIAP.93 SMAC and HtrA2 were identified as mitochondrial pro-apoptotic proteins by inhibiting IAP-binding motif domain; Kim et al. found a novel anti-apoptotic E3 Ub ligase, AREL1,4, which ubiquitinates and degrades SMAC, HtrA2 and ARTS (but not XAF1), only when they are released into the cytosol upon apoptotic stimulation.94
Additionally, caspase-3 and caspase-7 can be ubiquitinated by other E3 Ub ligase. CIBZ (ZBTB38 in humans) was found to be a novel substrate of caspase-3, and two caspase-3 recognition sites were identified, while knockdown of CIBZ in p53(−/−) mouse embryonic fibroblast cells also activated caspase-3 and cleavage of poly (ADP-ribose) polymerase, indicating that CIBZ plays an important role in the negative regulation of apoptosis in murine cells.95 Besides, caspase-3 can be stabilised by USP15, through which USP15 can control binding between procaspase-3 and SCF complex, thus resulting in the promotion of apoptosis.96 WWP2 was revealed to be significantly overexpressed in liver cancer tissues, its knockdown can induce G1 cell cycle arrest and apoptosis by significantly increasing the expression of caspase-7, caspase-8, Bax and decreasing Bcl-2.97
4 CANCER THERAPEUTIC APPROACHES TARGETING THE INTRINSIC APOPTOSIS PATHWAYS
4.1 Proteasome inhibitors
Most substrates labelled by Ub will be transported to the proteasome for degradation into small peptides, and abnormities in proteasome activity may affect normal protein degradation mechanisms. Cumulative data indicated that the survival of tumour cells depends on the activity of the proteasome, while the inhibition of the proteasome will tip the balance between pro-apoptotic proteins and anti-apoptotic proteins, facilitating in promoting the apoptosis of tumour cells.108 Therefore, proteasome inhibitors are exploited as effective targets to overcome chemotherapy sensitisation and inhibit tumour cell proliferation in the clinical treatment of cancer.109 Since Bortezomib (Velcade, PS341), the first proteasome inhibitor, was approved by the FDA for treating multiple myeloma in 2003, the development of anti-cancer drugs targeting the proteasome has been the most active research field, such as carfilzomib (Kyprolis) and ixazomib (MLN9708, Ninlaro).109
Bortezomib is a boronic acid-containing dipeptide that reversibly combines with the β5 subunit of the 20S proteasome and inhibits its chymotrypsin-like activity.110 On the one hand, bortezomib suppresses the UPS-mediated degradation of IκB, for another, it increases the levels of the pro-apoptotic protein NOXA.111, 112 Carfilzomib was proved to promote apoptosis via increasing NOXA levels, which leads to the activation of caspase-3 and caspase-7.113 Nevertheless, in spite of their effectiveness, proteasome inhibitors still have a limitation, including drug resistance, limited efficacy in solid tumours and hematotoxicity, preventing them from being developed as successful cancer treatments.114
4.2 Therapies targeting E3s
Proteasome inhibitors act on all substrate proteins degraded by the proteasome, while drugs targeting E3 can be targeted to stabilise a class of substrate proteins. This specific treatment may be more effective in improving efficacy while avoiding some non-specific side effects. The headmost approaches to target IAPs involve cancer therapeutics drugs that imitate the interaction of the endogenous IAP antagonist with IAPs, albeit with antisense approaches explored. Smac-mimetics (SMs) were originally designed to target and inhibit XIAP; however, subsequent research indicated that SMs bind with high affinity to cIAPs, and they can degrade cIAPs efficiently, not XIAP.115-117 Recently, ARTS-mimetics was demonstrated specifically combine with XIAP, promoting UPS-mediated degradation of both XIAP and Bcl-2, caspase activation and apoptosis.118 Dual downregulation of both major anti-apoptotic proteins, XIAP and Bcl-2, has been reported to cause enhanced apoptosis, increased sensitivity to chemotherapy and can overcome the resistance of cancer cells, ARTS-mimetics provide a promising novel platform for developing highly specific and potent anti-cancer drugs.118
E3 ligase MDM2 is another emerging target for developing cancer therapeutics strategies due to its inhibiting effect on p53-mediated apoptosis pathway.119, 120 Nutlin-3a, the first small molecule inhibitor targeting MDM2, is capable of simulating the spatial conformation of the key MDM2-binding site of p53 protein, and then replacing p53 to occupy the binding site, thereby hindering the degradation of p53 by ubiquitination modification and improving the intracellular level of p53.121, 122 Nutlin-3a shows a potent binding efficacy to MDM2 in many tumour models including chemoresistant neuroblastoma, acute myeloid leukaemia and multiple myeloma.123-125 Further optimisation of the nutlin-3 crystal structure promotes the discovery of better MDM2-p53 inhibitors, RG7112, which have exhibited greater activities in vitro.126 Nevertheless, RG7112 still shows many clinical adverse drug-related events including thromboycytopaenia and neutropaenia, rendering long-term treatment with RG7112 a major challenge.127
4.3 DUB Inhibitors
As mentioned, deregulated deubiquitination could lead to dysregulation of various critical events in apoptosis, thereby targeting DUBs may provide new strategies for anti-cancer therapy such as DUB Inhibitors (Table 2). While growing numbers of drugs have been developed to inhibit or antagonise DUBs, none of these has yet entered clinical trials. WP1130 can inhibit the activities of USP9X, USP5, USP14 and UCH37, downregulate the level of anti-apoptotic protein McL-1 and upregulate the level of tumour suppressor p53.128 Peterson et al. improved the drug-like properties of WP1130 and demonstrated that the novel compound EOAI3402143 dose-dependently inhibited Usp9x and Usp24 activity, increased tumour cell apoptosis and fully blocked or regressed myeloma tumours in mice.129 In addition, B-AP15 and its analogue VLX1570 specifically inhibit USP14 and UCHL5 without proteasomal activity inhibition.130 On the basis of DUBs substrate specificity, researchers can design different DUBs specific inhibitors for tumour therapy. It can be utilised in combination with traditional anti-cancer drugs, which is hopeful to promote the advancement of therapy sensitivity, reduce the generation of drug resistance, enhance the anti-tumour efficacy, and has a good clinical application prospect.
| DUBs inhibitors | Target | Therapeutic targets | Ref. |
|---|---|---|---|
| Proteasome inhibitors | |||
| Bortezomib | 20S proteasome | Multiple myeloma, mantle cell lymphoma, hepatocellular carcinoma and non-small-cell lung cancer | 110 |
| Carfizomib | 20S proteasome | Acute myeloid leukaemia, multiple myeloma | 113 |
| Ixazomib | 20S proteasome | Multiple myeloma | 131 |
| Marizomib | 20S proteasome | Glioblastoma | 132 |
| Oprozomib | 20S proteasome | Multiple myeloma | 133 |
| Therapies targeting E3s | |||
| Smac-mimetics | cIAPs | – | 117 |
| ARTS-mimetics | XIAP | – | 118 |
| Nutlin-3a | MDM2 | Chemo-resistant neuroblastoma, acute myeloid leukaemia and multiple myeloma | 123, 124 |
| RG7112 | MDM2 | Liposarcoma | 127 |
| DUB inhibitors | |||
| WP1130 | USP9X, USP5, USP24 | Mantle cell lymphoma | 128,,134, 135 |
| EOAI3402143 | USP 5, USP9X, USP24 | B-cell malignancies | 129 |
| VLX1570 | USP14, UCHL5 | Waldenstrom macroglobulinemia, multiple myeloma | 130 |
| F6 | USP2、USP7 | Lung adenocarcinoma | 136 |
| TCID | UCHL1, UCHL3 | Multiple myeloma | 137 |
| P5091 | USP7, USP47 | Multiple myeloma | 138, 139 |
| HBX 41,108 | USP7 | Colorectal carcinoma | 140 |
| b-AP15 | USP14, UCHL5 | Multiple myeloma, colorectal carcinoma, ovarian cancer | 137, 141 |
- Abbreviations: UCH, Ub C-terminal hydrolases.
5 DISCUSSION
Given that dwindled apoptosis promotes tumour development, induction of apoptosis for tumour cells has been a strategic concept for anti-cancer therapy for a long time. UPS play a significant role in modulating various phases of tumour cell apoptosis events, therefore targeting the UPS that may provide new inspiration and opportunities to operate cancer cell apoptosis. Here, we systematically summarised the mechanisms of Ub-specific enzymes especially E3 and DUBs-mediated regulation of cell-intrinsic apoptosis and their physiological association in cancer. Mitochondria positions at the heart of cellular metabolism and serves a pivotal role in apoptosis. Within the intrinsic mitochondrial pathway, the balance of anti- and pro-apoptotic Bcl-2 family members are modulated by E3s and DUBs, thus regulating their potential oncogenic behaviour in diverse cancers. Researchers have developed plenty of strategies targeting the UPS-mediated intrinsic apoptosis pathway, such as proteasome inhibitors, E3 ligase Inhibitors and DUB Inhibitors. However, only a few UPS inhibitors are still in use in clinical treatment, cancer drugs targeting the UPS at the present stage still exist limitations and challenges. For instance, bortezomib can produce peripheral neuropathy, fluid retention, fatigue, thrombocytopenia, nausea, vomiting and diarrhoea in MM patients, whereas B-AP15 can inhibit the growth of bortezomib resistant MM cells.137, 142 Therefore, researching new targets and combination therapy strategies rationally may be promising approaches. The most well-known feature of ubiquitination is breaking down proteins that are misfolded or outlived their usefulness. Given the mechanisms of Ub-mediated regulation of apoptosis and their physiological relevance, we hope that it will supply more therapeutic strategies for cancer.
[Correction added on January 17, 2023 after first online publication: the Author Contributions, Acknowledgments, Funding, Conflicts of Interest, Data Availability Statement and Ethical approval has been updated].
AUTHOR CONTRIBUTIONS
Y.L. and T.F. contributed to the preparation and collection of original literatures and figures and the writing. J.C. was responsible for the scientific quality and editing of manuscript.
ACKNOWLEDGEMENTS
Not applicable.
FUNDING
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Not applicable.




