Prostate cancer management with lifestyle intervention: From knowledge graph to Chatbot
Yalan Chen, Baivab Sinha, and Fei Ye contributed equally to this work.
Abstract
Background
Personal lifestyle is an important cause of prostate cancer (PCa), hence establishing a corresponding knowledge graph (KG) and a chatbot is a convenient way for preventing and assessing risks. The chatbot based on a KG of PCa-associated lifestyles will be helpful to PCa management, then save health care resources in the ageing society.
Results
Based on our established knowledge base, we define entities and corresponding relationships to construct the PCa-associated lifestyles KG for visualization by importing the triples into the Neo4j graph server. The dialogue system uses the Flask framework to determine the classification of questions through entity recognition and relationship extraction and later uses the query template to search the answers from the PCa-associated lifestyles KG. The PCa-associated lifestyles KG contains 11 types of entities and 14 types of relationships, the total number of nodes and links is 21 546 and 66 493, respectively. Also, the entity “Lifestyle”, “Paper”, “Baseline” and “Outcome” contain multiple attributes. The established chatbot can answer 12 types of basic questions and predict the probability of a certain lifestyle resulting in a certain PCa. The chatbot is available at http://sysbio.org.cn:5000/Pca/chatbot.
Conclusion
A chatbot based on PCa-associated lifestyles KG was constructed to help researchers, physicians or patients learn more about PCa lifestyle management interactively.
1 BACKGROUND
Despite the popular belief that many cancer cases result from an inherited genetic abnormality, 90% of malignancies are rooted in lifestyle and environmental exposure. Lifestyle medicine is a medical approach that uses evidence-based behavioural interventions to treat, manage and prevent modern diseases (mainly chronic, but potentially acute and infectious diseases) related to lifestyle.1-4 Research shows that over 80% of chronic conditions could be avoided through the adoption of a recommended healthy lifestyle, it was also reported that clinical events could be improved with positive lifestyle adoption.5
State-of-the-art artificial intelligence methods are increasingly leveraged in clinical predictive modelling to provide clinical decision support systems to physicians. Yet, these modern methods yield a limited understanding of the resulting predictions.6, 7 When we train artificial intelligence (AI) models with lots of parameters to which we apply transformations, we end up turning the entire process of pre-processing and model building into a black-box model that is very hard to interpret.8 However, in the medical domain, understanding applied models are essential, in particular, when informing clinical decision support. With the onset of Explainable AI, models could be developed with trustworthy and explainability. Using knowledge-guided models9, 10 and a well-received knowledge graph (KG) for AI-based clinical prediction is a step forward in this direction.11, 12
The KG belongs to a structured and displayable semantic network, initially known for improving the effectiveness of search engines.13 With the rise of artificial intelligence, KGs have been successfully used in risk assessment,14, 15 auxiliary diagnosis,16, 17 drug discovery,18, 19 and smart chatbot20, 21 in precision medicine. There exist several representatives and comprehensive KGs, such as IBM Watson,22 SNOMED-CT,23 and CmeKG,24 and so forth, most of which aimed to save time and vigour, alleviate the pressure of physicians, and improve the accuracy of diagnosis to some extent. By investigation, the existing professional medical KGs mainly focus on diseases,25 drugs,26 cells,27 literature,28, 29 proteins, genes, and organs.28 However, personal lifestyles such as diet, sleep, vitamins, and environment, which are critical triggers of diseases, such as prostate cancer (PCa), can be an inspiration for novel direction in this field.30
PCa is a heterogeneous disease with lethal and indolent phenotypes and is the most commonly diagnosed visceral cancer among men in most western countries.31 Many epidemiological and case-control studies disclosed that there is a great link between lifestyles and PCa, such as body weight, smoking, dietary factors, and also some other lifestyle-related diseases (hyperglycaemia and dyslipidemia).32 This makes the KG based on PCa lifestyle a perfect candidate for a novel direction in the field of lifestyle medicine. This KG will not only provide the rich pieces of evidence of PCa-lifestyle relations for experts and can serve a larger population, but also effectively speculate on the risks of single or integrated lifestyles for further suggestions.
At present, the construction of domain-specific KGs mainly relies on automated tools of natural language processing (NLP) or manual ways of extracting or merging structured information from electronic health records. Li et al. extracted knowledge from structured and semi-structured data and established a hepatocellular carcinoma-associated KG.25 Wang et al. established a KG for type 2 diabetes according to evidence reviewed by experts.33 Li et al. built a KG for knee osteoarthritis by training models to extract knowledge from electronic medical records.34 However, to our best knowledge, there exist no attempt to construct a professional KG for PCa-associated lifestyles.
Recently, we constructed the PCaLiStDB,35 a knowledge base (KB) for PCa lifestyles which was standardized further by lifestyle-wide association studies of PCa (PCa_LWAS).36 The PCaLiStDB consists of 300 qualified articles collected from PubMed, where 2290 single lifestyle factors and 856 combined lifestyle factors are extracted. In this article, we take advantage of the accuracy and standardization of the PCaLiStDB to innovatively build a KG, named PCa-associated lifestyles KG (PCalfst_KG), on the Neo4j platform. The resulting KG consists of 21 546 entities and 66 493 relationships. To make the query formation more intuitive, these graphs are further visualized using node.js and d3.js.37 In addition, to illustrate the practicality of the graph, we develop a primary chatbot based on the Flask framework to interactively assist users in understanding the potential risks that may arise by choosing a certain or a combination of lifestyles. This chatbot will not only be useful for medical professionals but also for common users (especially the elders) to analyze the effect of the association of different lifestyles for PCas.
2 METHODS
2.1 Data collection and processing
The PCaLiStDB which is standardized for PCa_LWAS is publicly available at http://www.sysbio.org.cn/pcalistdb/. In the PCaListDB database, there exists a total of 3024 lifestyles items comprising 394 protective items, 556 risk items, 45 uninfluential items, 52 ambivalent items and 1977 items that lack adequate literature support. These items are summarized and classified into three SQL tables. As observed in Figure 1, the PCaLiStDB consists of three SQL tables namely, pcalistdb_main.sql, pcalistdb_baseline.sql, and pcalistdb_outcome.sql. The pcalistdb_main.sql is composed of 300 records extracted from literature which include PMID, author, year, title, and study type. The baseline.sql table includes group number, index name, stratification, value, and notes. Finally, the outcomes.sql contains index_name, stratification, sample_size, PCa incidence, effect index, p-value and notes. Both of the tables contain 1000 records each. Further, the above-mentioned SQL files are converted into JavaScript Object Notation (JSON) format so that they can be read by python language's built-in functions.
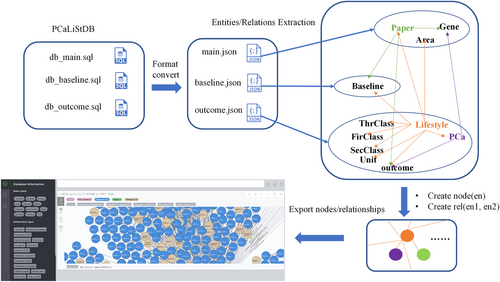
2.2 Entities/relations extraction
The graph space comprises an extensive set of triples, which include start or head entity (h), relationship (r), and end or tail entity (t), denoted as < h, r, t >. Each entity has its unique identifier, name, and corresponding properties, whereas each kind of relationship is designated by its name. We extract the entities and relationships from previously obtained JSON files. It is according to the existing and indirect key-to-key connections in the KB. As Figure 1 shows, we get entity “Lifestyle” (central entity), “PCa”, “Unit”, “Outcome”, and so forth from outcome.json; entity “Paper”, “Gene”, “Nation” from main.json; entity “Baseline” from baseline.json. Each orange arrow starting from “Lifestyle”, points to a unique entity, represents a kind of relationship. The same for the green arrow starting from “Paper” and a violet arrow starting from PCa. The first three columns of Tables 1 and 2 exhibit the definition of the distinct relationships between entities. For convenience, we define the name of the entity “Baseline” as “pbbase_id” and “Outcome” as “pcaoc_id”, respectively.
| Entity | Properties | Count |
|---|---|---|
| Lifestyle | factor_type, fenlei, index_name, inv_papers, level_class, name, paper_count, pca_type, unit. | 2290 |
| Paper | area, author, duration, gene, name, sample_size, study_type, title, year. | 300 |
| Baseline | group_number, index_name, name, notes, pmid, stratification, value. | 2570 |
| Outcome | aj_value, eaj, eunaj, index_name, name, notes, pcatype, pmid, stratification, unaj_value, unit. | 15 586 |
| PCa | no other specific property defined | 79 |
| Nation | no other specific property defined | 31 |
| Unit | no other specific property defined | 125 |
| Gene | no other specific property defined | 38 |
| FirClass | no other specific property defined | 11 |
| SecClass | no other specific property defined | 294 |
| ThrClass | no other specific property defined | 222 |
| Start entity | End entity | Relationship | Count | Proportion |
|---|---|---|---|---|
| Lifestyle | Paper | Related papers | 2985 | n:m |
| Lifestyle | PCa | Related leading PCas | 3564 | n:m |
| Lifestyle | Nation | Countries with lifestyle | 2807 | n:m |
| Lifestyle | Unit | Units of lifestyle | 1520 | n:m |
| Lifestyle | FirClass | First class of lifestyle | 2285 | n:1 |
| Lifestyle | SecClass | Second class of lifestyle | 2274 | n:1 |
| Lifestyle | ThrClass | Third class of lifestyle | 848 | n:1 |
| Lifestyle | Baseline | Baselines of lifestyle | 876 | 1:m |
| Lifestyle | Outcome | Outcomes of lifestyle | 15 575 | 1:m |
| Paper | Baseline | Baselines of paper | 2545 | 1:m |
| Paper | Outcome | Outcomes of paper | 15 583 | 1:m |
| Paper | Gene | Involved genes | 39 | 1:m |
| PCa | Outcome | Outcomes with PCa | 15 585 | 1:m |
| PCa | Gene | Genes with PCa | 7 | 1:m |
2.3 Knowledge import
We defined two functions, the first is to create an entity with a unique label as the only parameter along with its corresponding properties. Take the central entity “Lifestyle” as an example, the corresponding properties are: “name”, “fenlei”, “factor_type”, “level_class”, “index_name”, “inv_papers”, “paper_count”, “pca_type” and “unit”. The second function creates a relationship among three parameters: start entity label, end-entity label, and the relationship. Finally, entities and relationships generated from the above-mentioned functions are imported into the local Neo4j graph database. Every entity with its label is recorded. Finally, we acquire a total of 21 546 entities and 66 493 relationships between these entities.
As seen from Tables 1 and 2, 11 types of entities were obtained from the PCaLiStDB after the processing which includes lifestyle, paper, baseline, outcome, PCa, nation, unit, gene, FirClass, SecClass, and ThrClass. We also defined 14 categories of relationships between the entities. For a specific central entity, it contains multiple connected entities like “Paper”, “PCa”, “Nation”, “Baseline”, “Outcome” and “Unit”. However, a specific entity can be connected to multiple distinct entities. These multi connections are according to the proportion column of Tables 1 and 2. For example, there may be a scenario that a PCa can be associated with over one lifestyle, an article may involve multiple lifestyles, and so forth. It is worth noting that the map between entity types “Baseline” and “Lifestyle”, and entity type “Outcome” and “Lifestyle” is exclusive.
2.4 Performance
Textbox 1. Example query template
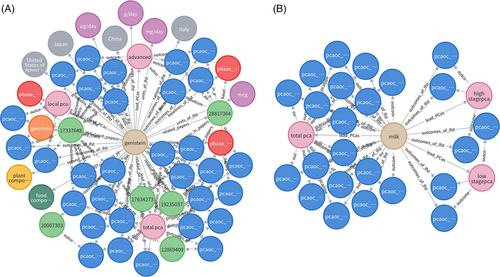
where p is all the accessible paths between n and m, n is the start (central) entity which has assigned the label, m represents the end entity, query represents the name of the start entity, rel_name represents the name of the relationship between them. As shown in Figure 2A, we found that the lifestyle “genistein” is present in six scientific articles in total, has four basic units including “g/day”, “mg/day”, “ug/day” and “msg”, appears in four countries or areas including “the USA”, “Japan”, “Italy” and “China”, may lead to the type of total, advanced and local PCa potentially, with three baselines and 31 outcomes. In addition, “genistein” belongs to the three-level classification with “food composition” (first), “plant compounds” (second) and, “genistein” (third).
Textbox 2. Query template
where p denotes the accessible path whose length is two, the template return all of the possible outcomes when the lifestyle leads to PCa to a certain degree. Figure 2A shows that lifestyle “genistein” may lead to 20 total PCas, eight advanced PCas and three local PCas.
Furthermore, Figure 2B shows that another lifestyle “milk” may lead to a total of 18 PCas, three high-stage PCas, and three low-stage PCas. From these observations, we can say that the “genistein” and “milk” are the important factors that may lead to prostate cancer.
2.5 Online visualisation
Based on the Neo4j graph database, we construct a front-end page of our user interface for intuitive interaction between the user and the system. Here, the user can input the name of a lifestyle to get the corresponding KG. It is convenient for users who are unfamiliar with Neo4j query language as they don't need to form the query on their own. For visual simplification, the corresponding retrieved map only keeps the direct relationships of the central node and the relationship between nodes of PCas and outcomes, which means the inessential relationships, such as links between literature and outcomes, literature and baselines, literature and genes, are omitted. For detailed information about nodes, such as lifestyles, literature, baselines and outcomes, users can inquire and browse specific information about the node in focus using the side property bar as per their need. In addition, the front-end page also provides the download function for several carrier data.
Textbox 3. Generated URL
localhost:3000/neo4j?query = URLEncode(query)
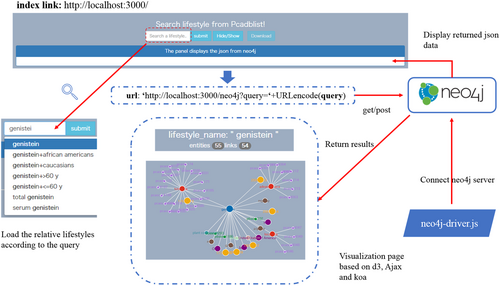
Textbox 4. Cypher query template
As the description above, the template first filters out the outcomes of several PCas caused by the requested lifestyle, thereafter it selects nodes that are directly connected to the central node. Finally, the resulting nodes, p and q, are combined for display. When the resulting data is returned, a dedicated panel will display it in the JSON format, which can be shown or hidden by a button click. The user can also download the JSON data using another button, named “download“.
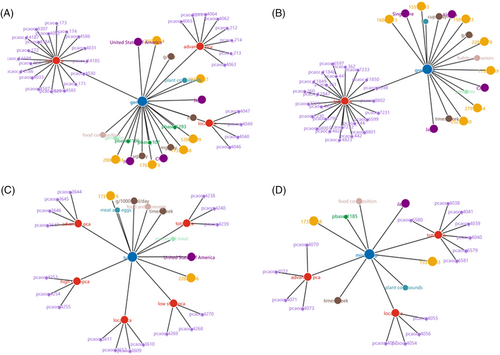
As shown in Figure 4, a visual mapping graph is automatically generated on a canvas element according to the returned JSON based on ajax and d3 technology. As observed from Figure 4, the title not only shows the name of the requested lifestyle but also demonstrates the other connected nodes and links. The interface contains six types of legends denoted by different colours scheme to distinguish distinct entities, and each node displays the name of the corresponding entity. Users will directly get all the information related to the lifestyle habits they want to query through the visualized page. When the mouse is focused on a certain node, its various properties and values will be displayed on the far right of the interface in detail, as shown in Figure 5, corresponding to the content of Table 1. While searching for a specific lifestyle, users can get all information about it on the front-end page. Additionally, users can also download the picture which is shows a related KG. A button is also provided that converts the SVG element to PNG format for convenient viewing after download.

3 RESULTS
3.1 From KG to chatbot
In this section, we established a chatbot based on a dialogue system that can answer basic questions about the lifestyle and how choosing a particular lifestyle can avoid the occurrence of prostate cancer. The system consists of two participants, one is the user and the other the bot. The framework of the dialogue system is divided into a front-end user interface to interact with the user and a back-end web server built on the Flask framework for query understanding and answer generation. The front interface not only captures the entered question and passes the question to the back-end through the get/post method, but also shows the generated answers based on the results inquired from the neo4j server. The back-end includes four different components, question classification, question parsing, answer searching, and answer generation, as presented in Figure 6.
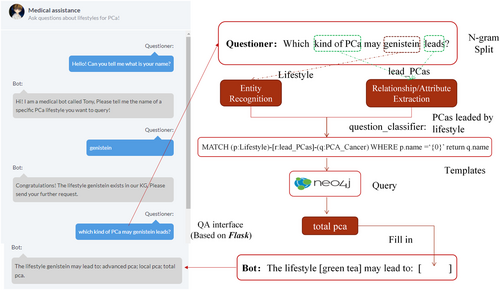
Before we recognize entities and extract relationships/attributes from an entered question, we should build a domain list consisting of a series of dictionaries, where the key is the entity type and the value is a set that includes all names of this entity from a text file which are stored in a single-line format and acquired from the PCalfst_KG. Additionally, we also construct a synonym list for keywords of a certain relationship or property type.
where punctuation is processed as an independent word. Here, N is the length of the question (N represents the words number in the question), n represents the size of the sliding window, “loop” means a time of traverse. The fdmat returns a Boolean, which aims to match tokens with the keywords in the domain list and the synonym list during the traversal to determine the classification of questions more precisely.
Next, at the stage of the question parsing, we select the corresponding Cypher template and send it as a query request to the neo4j server. Finally, if we retrieve the answer from the PCalfst_KG, the answer will be filled in the pre-designed reply template as a phrase slot. Finally, the generated reply will be presented to the user in the front-end user interface. Otherwise, the bot will give a hint that the answer cannot be retrieved from the PCalfst_KG.
3.2 Dialogue design for lifestyle-based PCa healthcare
| Q exm | Synonym | Classification | R exm |
|---|---|---|---|
| 1. Which papers are related to the lifestyle [genistein]? | survey, paper, investigation, research, report | Asks related papers | The PMIDs of related papers about genistein are 17634273, 19235037, … |
| 2. Can you give me brief information on paper whose PMID is [17634273]? | information, introduction, detailed information, specific information, core information, brief introduction, brief information |
Ask information (paper/baseline/outcome) |
The detailed information of the paper is as follows: Title: xxx Author: xxx … |
| 3. Can you give me brief information on baseline whose ID is [pbase_102]? | index_name: xxx; group_number: xxx; stratification: xxx; … | ||
| 4. Can you give me brief information on the outcome whose ID is [pcaoc_4584]? |
index_name: xxx; pcatype: xxx; eaj: xxx; aj_value: xxx; … |
||
| 5. How do we measure the lifestyle [genistein]? | measurement, unit, measure, dosage | Asks units | The corresponding units of genistein are g/day, mg/day, … |
| 6. Where the lifestyle [genistein] may appear? | where, area, country, nation, region, location, appear | Asks geographical areas | From KG, we find that the genistein appears in Japan, China, Italy, … |
| 7. Can you give the involved baselines of lifestyle [genistein]? | – | asks baselines of lifestyle | The possible baselines of genistein are pbase_102, … |
| 8. What is the influence factor of lifestyle [genistein]? | – | Asks influencing factor | The influence factor of genistein is “No statistical significance factor”, “Protective factor; impact level:Strong”. |
| 9. Which class level does the lifestyle [genistein] belong to? | kind, class, type, classification, belong | Asks class level | The genistein belongs to class: food composition, plant compounds, genistein. |
| 10. Lifestyle [genistein] can lead to which kind of PCas? | PCa, illness, sickness, disease, pathema, prostate cancer, prostatic carcinoma, CRPC, prostatic cancer, cancer | PCas led by lifestyle | The genistein may lead to total/local/advanced PCa. |
| 11. When lifestyle [genistein] leads [total PCa], please give possible outcomes. | – | Asks about the outcomes of PCas | The total PCa may bring about outcomes: pcaoc_4584,…. |
| 12. How many genes may [advance PCa] be associated with? | – | Asks about associated genes | The advance PCa is associated with genes such as xxx. |
Otherwise, we continue the Q&R process of the input lifestyle. As shown in the question column (Q exm) of Table 3, the entities are circled with brackets, whereas the keywords associated with relationship or attributes are marked with the solid line; in the reply column (R exm), answers retrieved from the KG are marked by the dotted line.
3.3 The realization PCa healthcare chatbot
Textbox 5. Resulting Cypher template
where the numerator is the count(m), the denominator is the number of lifestyles that may lead to the , without the conditional statements in our template.
- Q: I have a habit of taking the genistein, which kind of PCa may it lead to?
- A: Your habit may lead to PCa1, PCa2, …
- Q: Can you predict the risk rate of getting advanced PCa under taking the genistein?
- A: The risk rate is: (PCa1: rt1), (PCa2: rt2), …
4 DISCUSSION
4.1 Principal novelty and potential applications
In this article, we established the novel KG called PCalfst_KG associated different lifestyle habits to prostate cancers. To the best of our knowledge, this is the first KG in this domain. The graph consists of 21 546 entities and 66 493 relationships. For intuitive visualization of our KG, PCalfst_KG, we developed a user interface using d3 and node.js web technology to facilitate the query formation of the users who are not familiar with the neo4j query language. We established a chatbot based on dialogue system on the Flask Framework. The chatbot can answer 12 basic questions about a certain PCa lifestyle.
4.2 Resources
The KG and the chatbot are available online at http://sysbio.org.cn:3000/. And http://sysbio.org.cn:5000/Pca/chatbot respectively. We also release the demo source code at https://github.com/rshsm/Impact-of-Lifestyle-on-PCas-from-Knowledge-Graph-to-Chatbot for other researchers’ use.
4.3 Limitations and future work
The established KG is not extensive, with fewer entities and relationships, resulting in a lesser robust graph. Moreover, the nodes associated with the central lifestyle node are also insufficient. The above-mentioned problems not only result in the lack of diversity in our graph but also limit the random questions that the chatbot can support. Also, since the PCalfst_KG is the first established KG about PCa habits, we have not found effective indicators to evaluate the performance of the current KG, including scalability, accuracy, coverage, and response time.
The chatbot only performs simple entity recognition and relationship/attribute extraction of the requested questions, and the answer of each question is set unique by default. As the number of entities grows further, the retrieval process of entries that rely on the domain dictionary will be more time-consuming. Additionally, the chatbot is based on query templates rather than a deep learning or machine learning model, which leads to the inflexibility of the system. Hence, the evaluations of current state-of-art NLP models are not applicable for our system.
In the future, we should first establish and strengthen the ontology of the PCa lifestyles to achieve effective entity disambiguation and entity alignment. In order to periodically upgrade the scale of the PCalfst_KG, it is necessary to train the named entity recognition model with BiLSTM-CRF39-41 or Tree-LSTM40, 42 supervised on existing entity-tag data from newly published medical literature or case reports for entities recognition and relations extraction.
To solve the problem of inflexibility and lack of diverse corpus for the chatbot, we will combine the template-based methods and deep learning methods. That mean pre-defining standard templates for Q&A pairs which are considered synonyms, filling the word slots according to the search results to get batches of Q&A instances, then generating a corpus of Q&A pairs based on the method of deep generative models combined with a threshold such as Generative Adversarial Networks43 or Variational Autoencoder.44-46 By calculating the similarities between the requested input questions and questions available in the Q&A system, alternative answers can be given. Furthermore, the generated corpus can train the seq2seq47, 48 or the chatbot directly, which may reduce the retrieval time as compared to the previous methods. Finally, the chatbot should also support the voice or image interaction with the users and we could also add multi-round user interaction to clarify the request.
5 CONCLUSION
The PCa associated lifestyle KB was transformed into a professional KG and conveniently visualized. We have initially constructed a chatbot based on the KGs, which is helpful to researchers, physicians or even patients for the personalized management of PCa lifestyles and assessment of PCa risks. To our best knowledge, this is the first chatbot in the cancer healthcare field to apply lifestyle KG for cancer risk assessment and prevention, the future extension and updating of this tool will include case studies and recommendation system, then make this knowledge-guided healthcare paradigm practical in the daily life.
[Correction added on January 17, 2023 after first online publication: the Author Contributions, Acknowledgments, Funding, Conflicts of Interest, Data Availability Statement and Ethical approval has been updated].
ACKNOWLEDGEMENTS
Not applicable.
FUNDING
National Natural Science Foundation of China, Grant/Award Numbers: 32070671, 82102186; The regional innovation cooperation between Sichuan and Guangxi Provinces, Grant/Award Number: 2020YFQ0019.
CONFLICT OF INTEREST
The authors declare no conflict of interest. The paper was handled by editors and has undergone a rigorous peer-review process. Dr. Shen was not involved in the journal's review of/or decisions related to this manuscript.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Not applicable.




