Deubiquitinating enzymes for targeted cancer therapy
Graphical Abstract
In this commentary, we briefly introduce the ubiquitin-proteasome system and the potential targets for drug discovery in this system. We then outline the classification and function of deubiquitinating enzymes (DUBs) and discuss the molecular mechanism by which OTUB1 regulates the deubiquitination and function of cellular communication network factor 6, a specific tumour suppressor in breast cancer. Finally, we summarise the strategies for the regulation of DUBs at different levels.
1 THE UBIQUITIN-PROTEASOME SYSTEM (UPS) AND DRUG DISCOVERY
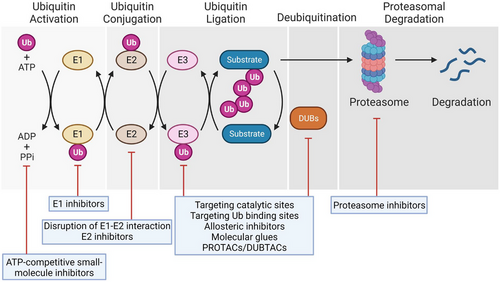
Proteostasis is essential for the maintenance of normal physiological functions, and its disruption leads to a variety of diseases, including cancers and neurodegenerative diseases.1 In eukaryotic cells, there are two major pathways for protein degradation: the UPS and the autophagy-lysosome pathway.2 In the UPS, proteins are covalently tagged with ubiquitin molecules through a cascade of multiple enzymatic reactions and then recognised by the 26S proteasome for degradation if specific ubiquitin chain linkages are formed on the targeting proteins. The conjugated ubiquitin on the substrates could also be removed by deubiquitinating enzymes (DUBs), thereby resulting in the alteration of their stability, localisation and protein–protein interaction as well as their biological functions.
The enzymes and key components in the UPS could be utilised for the targeted drug discovery (Figure 1). Adenosine triphosphate (ATP), activating enzymes E1s, conjugating enzymes E2s, ubiquitin ligases E3s, DUBs and the 26S proteasome could be potential targets for therapeutic treatment because their inhibition could prevent protein ubiquitination and/or subsequent degradation, thereby alleviating the abnormal growth of cancer cells. In fact, bortezomib, a proteasome inhibitor, is the first Food and Drug Administration (FDA)-approved drug that targets the UPS for the treatment of haematological malignancies.3 TAK-243 potently and selectively inhibits the E1 activating enzyme UBA1 by forming the TAK-243–ubiquitin adduct, causing the reduction in cellular conjugated ubiquitin and the death of cancer cells.4 CC0651 is an allosteric inhibitor that specifically targets the E2 conjugating enzyme Cdc34, leading to the accumulation of two downstream targets cyclin E and p27 and the inhibition of cancer cell proliferation.5 Several compounds including Nutlin-3 can disrupt the interaction between the E3 ubiquitin ligase Murine double minute 2 (MDM2) and its substrate p53, a well-known tumour suppressor, by binding to the p53-binding pocket in MDM2, resulting in the stabilisation of p53 and thereby suppressing the growth of tumour cells.6 Ubiquitin carboxyl-terminal hydrolase 7 (USP7, also called HAUSP) could remove the conjugated ubiquitin from MDM2 and thus promote the ubiquitination and degradation of p53. Therefore, inhibition of the catalytic activity of USP7 could indirectly elevate the p53 protein level, leading to the augmented apoptosis of cancer cells.7, 8 In contrast, in other cases, activation or upregulation of DUBs could remove the conjugated ubiquitin on substrates and thus directly prevent their degradation mediated by the 26S proteasome.
2 FUNCTION OF OTUB1 IN THE REGULATION OF CELLULAR COMMUNICATION NETWORK FACTOR 6 (CCN6) UBIQUITINATION AND BREAST CANCER GROWTH
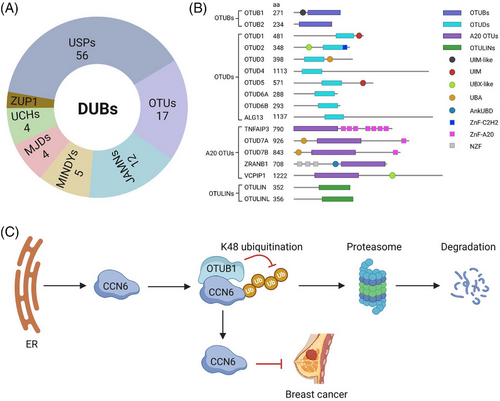
The canonical function of DUBs is to remove the conjugated ubiquitin from protein substrates. If the removed ubiquitin on the substrates is deemed to proteasome degradation, DUBs enhance the stability of the substrates, increase their activity and promote their biological functions. Therefore, dysregulation of DUBs leads to multiple diseases.9 Based on the sequence conservation of the functional domains, DUBs are categorised into seven families,10, 11 including Ubiquitin-specific protease (USP), ovarian tumour domain-containing proteases (OTU), JAB1/MPN/MOV34 metalloenzymes, motif-interacting with ubiquitin-containing novel DUB family, Machado–Josephin domain-containing proteases, ubiquitin carboxy-terminal hydrolases and zinc finger-containing ubiquitin peptidase 1 (Figure 2A). OTU family is one of the major types of DUBs, which is subclassified into OTUB, OTUD, A20 and OTULIN (Figure 2B). Among them, OTUB consists of two members, OTUB1 and OTUB2. OTUB1 regulates the deubiquitination and biological functions of several substrates, including ERα, p53, Ras and Snail (Table 1). It has been disclosed that CCN6 (also known as WISP3) is a specific tumour suppressor for breast cancer and can inhibit the growth, invasion and metastasis of cancer cells.12 However, it is unknown whether this protein could be regulated by the UPS or not.
| # | Cancer type | Substrates | Biological function | Inhibitors | References |
|---|---|---|---|---|---|
| 1 | Bladder cancer | ATF6 | Stabilising ATF6 and inhibiting proliferation, viability and migration | / | 15 |
| E2F1 | Leading to E2F1 deubiquitination, promoting proliferation, migration, invasion and inhibiting cell apoptosis | / | 27 | ||
| 2 | Breast cancer | MYC | Blocking MYC degradation, promoting breast tumourigenesis | / | |
| 3 | Breast cancer | Cellular communication network factor 6 (CCN6) | Stabilising CCN6, inhibiting the migration, proliferation and viability | / | 13 |
| 4 |
Breast cancer, renal cell carcinoma, prostate cancer |
FOXM1 |
Stabilising FOXM1 and enhancing the epirubicin resistance of breast cancer; Stabilising FOXM1, promoting RCC cell viability, migration, invasion and proliferation; Stabilising FOXM1 and promoting metastasis of prostate cancer |
Atorvastatin | 28-30 |
| 5 | Endometrial cancer | ERα | Stabilising ERα, affecting ERα-dependent transcription | / | 31 |
| Aesophageal cancer | RhoA | Stabilising RhoA, promoting tumourigenesis and metastatic activity | / | 32 | |
| 6 | Aesophageal cancer | Snail | Stabilising Snail and facilitating metastasis | / | 33 |
| 7 | HEK293 | p-small mothers against decapentaplegic homolog 2/3 (SMAD2/3) | Inhibiting phospho-SMAD2/3 ubiquitination | / | 34 |
| 8 | HEK293T | RNF168 | Suppressing RNF168-dependent polyubiquitination and DNA damage response | KU55933 | 35 |
| 9 | HEK293T | DEP domain-containing mTOR-interacting protein (DEPTOR) | Stabilising DEPTOR, suppressing mTOR complexes 1 (mTORC1) activation | / | 36 |
| 10 | Lung cancer | Ras | Inhibiting Ras monoubiquitination, enhancing tumourigenic growth | Analogues of SJB2-043 | 37,38 |
| 11 | Lung cancer | p53 | Inhibiting p53 ubiquitination, promoting DNA damage response and inducing apoptosis | Analogues of SJB2-043 | 31,39 |
| 12 | Multiple myloma (MM) | C-Maf | Abrogating c-Maf polyubiquitination, promoting MM cell survival and MM tumour growth |
Lanatoside C Nanchangmycin |
39, 40 |
| 13 | Non-small cell lung cancer | p-YAP | Activating OTUB1/Hippo signalling pathway, promoting proliferation, migration, invasion and sphere formation | / | 41 |
| 14 | Prostate cancer | Cyclin E1 | Stabilising cyclin E1, promoting cancer cell proliferation and progression | RO-3306 | 42 |
A paper published recently in Clinical and Translational Medicine by Zhao et al.13 revealed that the degradation of CCN6 is regulated by the proteasome. Through a DUB screening, they unveiled that OTUB1 expression significantly increases the cellular CCN6 protein level but not the secreted CCN6 in breast cancer cells. However, its mRNA level is not altered by OTUB1. In line with this, OTUB1 knockout decreased the CCN6 protein level. Mechanistically, OTUB1 reduces the K48-linked polyubiquitin chains on CCN6 and thus inhibits its degradation through the endoplasmic reticulum-associated protein degradation process (Figure 2C). Deletion of the UBA and linker domains but not the UBA domain alone in OTUB1 completely abolishes the OTUB1-CCN6 interaction, suggesting that the linker domain in OTUB1 is necessary for its binding to CCN6. Molecular docking and biochemical experiments further discovered that the noncatalytic residue D88 but not the catalytic cysteine C91 is required for the reduction of the CCN6 ubiquitination, indicating that OTUB1 inhibits CCN6 ubiquitination through the non-canonical manner by obstructing the E2/E3-dependent transfer of ubiquitin molecules. This is further supported by the fact that the full length and the C91S OTUB1 but not the D88A mutation stabilise CCN6.
Having elucidated the molecular mechanism by which OTUB1 reduces the ubiquitination and degradation of CCN6, the authors further demonstrated that OTUB1 deficiency enhances the migration, proliferation and viability of breast cancer cells, while replenishment of CCN6 remarkably hampers these processes. The xenograft mouse models subcutaneously injected with OTUB1 knockout or OTUB1 knockout and CCN6-expressing breast cancer cells confirmed the roles of OTUB1 and CCN6 in mediating the growth of breast cancer cells and the regulation of OTUB1 on CCN6. Using clinical samples, the authors further uncovered the positive correlation between OTUB1 and CCN6 at the protein level in breast cancer tumour tissues.13
3 PERSPECTIVES ON THE CANCER THERAPY TARGETING DUBs
A wealth of studies demonstrated that DUBs could regulate the proliferation, migration and invasion of cancer cells by modulating the ubiquitination, degradation and thus stability of tumour suppressors or oncoproteins.14 Therefore, these DUBs could be potential therapeutic targets for cancer treatment. The functions of DUBs may be cancer-type-specific. For example, OTUB1 could function as an oncoprotein and promote the growth of multiple cancer cells, including bladder cancer,15 colorectal cancer,16 oesophageal cancer,17 lung cancer18 and multiple myeloma.19 It has been discovered that OTUB1 can regulate the ubiquitination and degradation of multiple targets in cancer cells (Table 1). OTUB1 contributes to breast tumourigenesis by stabilising an oncoprotein MYC.20 However, OTUB1 also acts as a tumour suppressor because it increases the stability and transcriptional activity of p53.21 However, these two substrates are not specific for breast cancer. In the above-discussed paper, Zhao et al.13 discovered a new target for OTUB1, CCN6, a potent tumour suppressor specific for breast cancer. Since multiple substrates with completely different functions in various cancer cells have been discovered for OTUB1, caution should be taken when OTUB1 is utilised as a clinical target for cancer therapy.
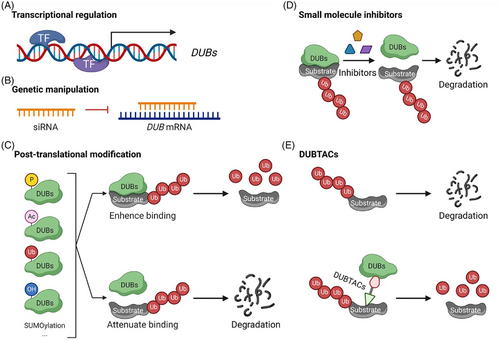
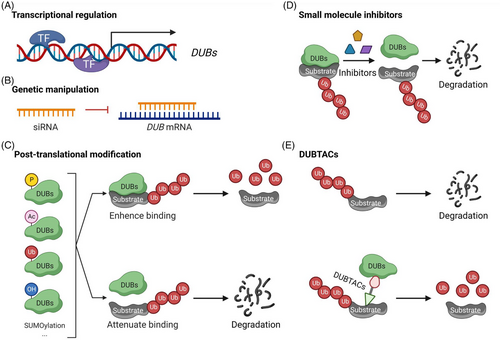
Several common strategies may be used to manipulate the expression and activities of DUBs. First, alteration in transcription factors (TFs) or transcription regulators could modulate the expression of a DUB gene of interest (Figure 3A). Second, transfection or delivery of siRNA to cells could genetically knock down a DUB mRNA and the expression of its encoded protein (Figure 3B). Third, DUBs could also be regulated by a variety of post-translational modifications (PTMs) such as phosphorylation, acetylation, hydroxylation, oxidation, ubiquitination, SUMOylation and proteolysis (Figure 3C).10, 22, 23 These PTMs could alter the stability, localisation, activity and protein–protein interaction of DUBs. For example, phosphorylation and acetylation can either enhance or attenuate the deubiquitinating activities of DUBs by modulating their binding to substrates. Fourth, small molecule inhibitors could be developed to block the enzymatic activity of DUBs (Figure 3D). Inhibitors for USP7 and other DUBs can attenuate the proliferation of cancer cells.24 Moreover, to prevent the degradation of proteins through the UPS, deubiquitinase-targeting chimeras (DUBTACs; Figure 3E) were developed to remove the conjugated ubiquitin on protein substrates and to thereby enhance protein stability.25 TFs can be stabilised by TF-DUBTACs by removing the conjugated ubiquitin and diminishing their degradation. This strategy has been used to stabilise forkhead box class O3A (FOXO3A), p53 and interferon regulatory (IRF3).26 These DUBTACs utilise OTUB1 to deubiquitinate the corresponding TFs for their stabilisation. Additional strategies will be developed in the future to explore other strategies targeting DUBs for cancer therapy.
AUTHOR CONTRIBUTIONS
Yue Li and Xiaohui Wang. writing-original draft; Xiaohui Wang. writing-review and editing.
ACKNOWLEDGEMENTS
This work was supported by the interdisciplinary seed funding in medical science from Suzhou Medical College of Soochow University (MP13207823) and a project funded by the Priority Academic Program Development (PAPD) of the Jiangsu Higher Education Institutions of China.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.