An efficient large-scale whole-genome sequencing analyses practice with an average daily analysis of 100Tbp: ZBOLT
Zhichao Li, Yinlong Xie, and Wenjun Zeng contributed equally to this work.
Abstract
Background
With the advancement of whole-genome sequencing (WGS) technology, massively parallel sequencing (MPS) remains the mainstream due to its accuracy, low cost, and high throughput. The development of the analytical pipeline corresponding to MPS has always been of great importance. Increasingly large population genomics studies, as a specific type of big data research, pose new challenges for analysis solutions.
Results
Here, we introduce ZBOLT, a comprehensive analysis system that incorporates both software and hardware advancements, making it an appropriate choice for large-scale population genomic studies that require extensive data processing. In this study, we first evaluate ZBOLT's calling accuracy using the Genome in a Bottle (GIAB) benchmark dataset. Then we apply ZBOLT to a large-scale population genomics study with 5,616 high sequencing depth samples totaling 1.16Pbp (base pair). As the results show, ZBOLT demonstrates exceptional efficiency and low energy consumption, processing 100Tbp per day and using 1kWh per 100Gbp sequenced sample.
Conclusion
This research serves as a valuable reference for analyzing sequencing data from large population cohorts and underscores the significant potential of ZBOLT in large-scale population genomics studies.
1 INTRODUCTION
Since the launch of the Human Genome Project (HGP), DNA sequencing technology has been developed rapidly.1 Massively parallel sequencing (MPS) technologies, such as sequencing by synthesis (SBS)2 and DNA nanoballs (DNBs) sequencing3 have been widely used in recent years due to their good performance in terms of accuracy, throughput, speed, and cost.4, 5 As sequencing costs decrease, an increasing number of large-scale population genomics programs are being conducted to investigate the genetic variation among different populations, uncover diverse genetic backgrounds and support precision medicine initiatives.6 Examples of such programs include UK Biobank,7, 8 TOPMed9 and “All of Us” of NIH.10 However, challenges remain in analysis efficiency, power consumption, and the cost of supporting infrastructure when performing analyses for larger population genomic studies.11
To date, the majority of MPS analysis pipelines have been primarily designed to enhance individual analysis performance. The first widely recognized MPS analysis pipeline, the Genome Analysis ToolKit (GATK) Best Practices,12 was released by the Broad Institute in 2013. This workflow includes key software such as BWA13 and GATK,14, 15 utilizing nontrivial statistical modules like logistic regression, Hidden Markov Model, naive Bayes classification and Gaussian mixture model for variants calling.15
Since then, the demand for timely acute diagnostics in the translational application of disease and genomic studies of an increasing number of research populations has driven the development of more efficient and accurate MPS analysis pipelines. Representative pipelines include Edico DRAGEN,16 Sentieon DNASeq17 and DeepVariant.18 Edico DRAGEN accelerates analyses primarily through field-programmable gate arrays (FPGAs).16 Sentieon DNAseq follows GATK Best Practices by rewriting codes for optimizing analysis efficiency without modifying statistical modules.17 With the advancement of deep learning, DeepVariant has been developed, utilizing deep convolutional neural networks to call variants.18 This approach has demonstrated higher accuracy and efficiency in calling SNPs and InDels compared to GATK Best Practices.18-20
Population genomics studies, in contrast to individual studies, require the analysis and processing petabyte-scale data, presenting challenges in terms of accuracy, efficiency, and affordability.21, 22 Several existing pipelines heavily reliant on graphics processing unit (GPU) performance,23, 24 further complicating these challenges. In response to these issues, we explored ZBOLT, a high-performance resequencing analysis system. ZBOLT is designed to handle large-scale studies such as our newborn genomics and molecular epidemiology study consisting of 5,616 high-depth sequencing samples.
ZBOLT incorporates the MegaBOLT pipeline and is optimized for population analysis for WGS, including germline and somatic mutation calling, whole exome sequencing (WES), and targeted region sequencing. Adhering strictly to GATK Best Practices,12 ZBOLT leverages a heterogeneous computing system, employing central processing unit (CPU) and FPGA resources, dynamic multi-task scheduling, and hardware configuration support for accelerated analysis. ZBOLT utilizes a customized scheduling algorithm designed specifically to optimize multi-node computing scheduling within heterogeneous environments. The acceleration of the process from FASTQ to variant call format (VCF) results is achieved through data segmentation, optimization of compression/decompression algorithms, and streamlined calculation models. Moreover, with ZBOLT's on-premise analysis system, we prioritize enhanced data security and privacy, providing a distinct advantage over traditional cloud computing solutions.
Using ZBOLT, we completed quality control, alignment, and variant calling of 5,616 high-depth WGS data (approximately 1.16P base pairs) in 11.6 days. ZBOLT's efficient use of energy and cabinet space alleviates the efficiency and cost challenges associated with large-scale genomics data, particularly in the context of high-depth WGS analysis.
2 MATERIALS AND METHODS
2.1 Accuracy evaluation using benchmark datasets
To assess the accuracy of the ZBOLT system, we conducted three replications of WGS analysis experiments using benchmark datasets from five individuals (HG001-HG005). These datasets consisted of five paired published FASTQ25 with a coverage depth of approximately 30X per sample, sequenced by MGISEQ2000 and stored in GIAB26 Github repository (Table S1). The reference genome used for the analysis experiment and evaluation was GRCh38. Corresponding benchmark (high-confidence) variant calls and regions were stored in VCF27 and BED (Browser Extensible Data format) files marked NIST3.3.2 and GRCh38 (https://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/release/*/NISTv3.3.2/GRCh38/). The analysis results of the same samples running GATK (GATK3.8) best practices applied to the same samples were used as controls during the evaluation.
2.2 Experimental preparation in the newborn genomics study
To evaluate the performance of ZBOLT in large-scale data analysis, we collected samples from a newborn genomics study. In this study, we obtained 5,616 samples and extracted genomic DNA from newborn blood spots using the MagPure Tissue&Blood DNA LQ Kit (Magen, Guangzhou, China). After DNA extraction, Qubit 3.0 fluorometer (Life Technologies, Paisley, UK) was used to measure the DNA concentration and 2% agarose gel electrophoresis was used to assess DNA fragment integrity. Following the MGI's (MGI, Shenzhen, China) instructions, we proceeded to library construction and sequencing on the DIPSEQ platform of MGI (MGI, Shenzhen, China) with 100-bp paired-end reads. After sequencing, reads were stored in FASTQ25 format for subsequent bioinformatics analysis with the ZBOLT system.
2.3 Accuracy evaluation process
We used the ZBOLT WGS analysis pipeline to call variants for the five public FASTQ datasets (HG001-HG005). Subsequently, we compared the SNP and InDel VCF27 results with GIAB benchmark variants in the high-confidence region using RTG vcfeval.28 Precision, sensitivity, and F-measure were calculated to evaluate the accuracy of the ZBOLT system. Additionally, we assessed the consistency of the ZBOLT system by comparing its analysis results with those obtained using the widely recognized GATK Best Practice pipeline.
2.4 ZBOLT system with MegaBOLT pipeline
The ZBOLT workflow, as illustrated in Figure 1, utilizes a heterogeneous computing system of CPU and FPGA to accelerate WGS analysis from FASTQ25 input of sequencing data to BAM28 output of alignment result, as well as VCF27 / GVCF (Genomic Variant Call Format) output of variant calling result. ZBOLT employs a parallel computing architecture for FPGAs and processes data in smaller granules to enhance efficiency. A multi-task scheduling system and parallelized computing architecture improve the efficiency of WGS/WES analysis modules, including SOAPnuke,29 BWA,13 GATK14 and MuTect2.30 The documentation for ZBOLT is available on the official software repository at (https://en.mgi-tech.com/products/software_info/6/).
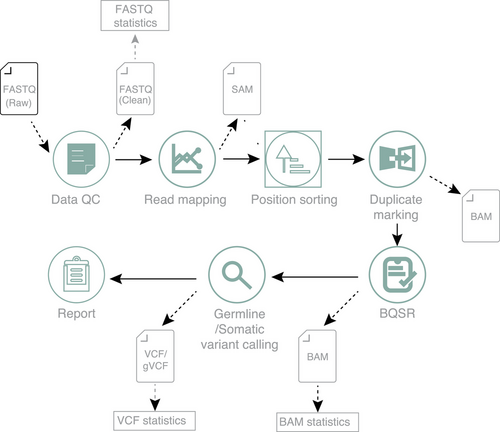
2.5 Population genomics study evaluation
In the newborn population genomics study, we calculated the pain points during the process of large population genomics dataset, including cabinet space, analysis time, power consumption, data transfer rate and data storage performance. To examine the reliability of analyses in large population studies, we finally performed quality control on the results of population analyses.
3 RESULTS
3.1 ZBOLT system efficiency and performance
A ping-pong buffer31 structure is adopted in the data organization of FPGA global memory (FPGA GM) to achieve high I/O efficiency. Data segmentation, compression/decompression algorithm optimization, calculation model simplification, index optimization contribute to the acceleration of the process from FASTQ to VCF results.
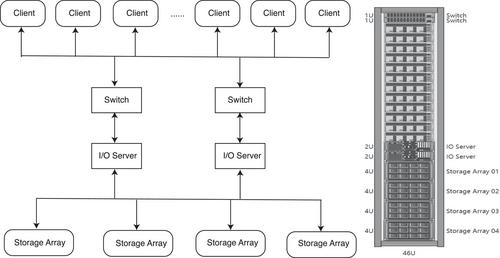
ZBOLT is supported by ZTRON (Figure 2), a specific genomics data management system designed to cater to the specific needs of multi-computing node collaboration and large file processing in population genomics research. Table 1 presents a comparison of the performance of ZTRON and general products in terms of capacity, density, data transmission rate and data protection.
| Regular products | ZTRON data management system | |||
|---|---|---|---|---|
| Capacity | 1# | 2# | 3# | |
| Density | 36 disks per 4U chassis | 60 disks per 4U chassis | 84 disks per 5U chassis | 106 disks per 4U chassis |
| Single node IO performance | 2.8 GB/s | 4 GB/s | 7 GB/s | 10 GB/s |
| Data transmission | Single 10/25 Gb/s | Multiple 100 Gb/s | ||
| Data protection | EC | RAID | RAID | RAID6 and hot disk redundancy backup |
A RAID0 (Redundant Array of Independent Disks)32 configuration, consisting of two SSDs (Solid State Drives), enhances the I/O between ZBOLT and the storage module, caches all intermediate files in the calculation process and improves the system robustness. The customized scheduling algorithm facilitates multi-node computing scheduling in heterogeneous environments (Figure 3), intelligently allocating tasks to different nodes based on their resource utilization.
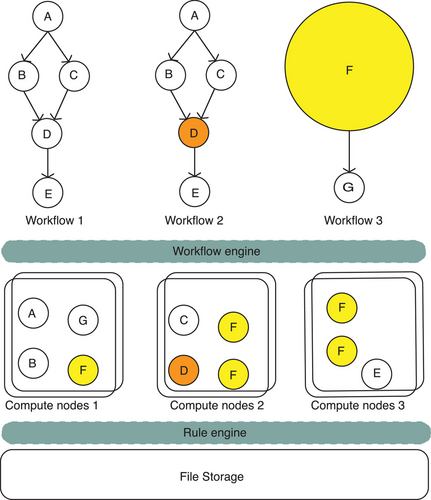
A proprietary file system and rule engine for data management enable the dynamic management of genetic data throughout the entire cycle, improving storage utilization rates. Cyclic redundancy check is employed to guarantee both data integrity and efficient data transmission speed during the data transfer process. The portable workflow description language and visual interface design allow for convenient editing of the analysis process when facing various research demands, such as somatic mutation analysis. The performance of the ZBOLT system is supported by multiple hardware and software configurations.
3.2 ZBOLT performance in individual analysis
We evaluated the accuracy of ZBOLT pipeline by comparing the called variants from ZBOLT VCF27 outputs with the GIAB benchmark as baseline variants. The GATK Best Practices outputs were used as control, which analyzed the same sample with the same parameters. The F-measure was used to evaluate the accuracy of SNP and INDEL calling, as shown in Table 2. The average F-measure in SNP calling evaluation is 99.65%, and for INDEL calling evaluation is 99.098%. This indicates that the ZBOLT process had high accuracy for both SNP and INDEL calling. The results of the three replicates were also highly consistent, indicating the stability of the ZBOLT system (Table S2).
| Sample & PipelineHG001-HG005 | Metrics | |||||
|---|---|---|---|---|---|---|
| SNP | INDEL | |||||
| Precision | Sensitivity | F-measure | Precision | Sensitivity | F-measure | |
| HG001.GATK | 99.55% | 99.82% | 99.68% | 99.11% | 98.99% | 99.05% |
| HG001.ZBOLT | 99.55% | 99.82% | 99.68% | 99.11% | 98.98% | 99.05% |
| HG002.GATK | 99.46% | 99.82% | 99.64% | 99.29% | 99.40% | 99.34% |
| HG002.ZBOLT | 99.46% | 99.82% | 99.64% | 99.29% | 99.40% | 99.35% |
| HG003.GATK | 99.49% | 99.82% | 99.66% | 99.11% | 99.04% | 99.08% |
| HG003.ZBOLT | 99.49% | 99.82% | 99.66% | 99.11% | 99.04% | 99.08% |
| HG004.GATK | 99.52% | 99.76% | 99.64% | 99.04% | 98.73% | 98.89% |
| HG004.ZBOLT | 99.52% | 99.76% | 99.64% | 99.05% | 98.73% | 98.89% |
| HG005.GATK | 99.43% | 99.83% | 99.63% | 98.95% | 99.28% | 99.12% |
| HG005.ZBOLT | 99.43% | 99.83% | 99.63% | 98.95% | 99.29% | 99.12% |
Moreover, the accuracy of the ZBOLT system was found to be comparable to that of GATK Best Practices when using the same inputs and parameters. The results of the GATK Best Practices analysis pipeline demonstrated that the average F-measure for SNP calling evaluation is 99.65%, while it is 99.097% for INDEL calling evaluation (Table S2).
An evaluation of the concordance between GATK Best Practices and ZBOLT was also conducted. As depicted in Table 3, the average F-measure in SNP calling was 99.99% and for INDEL calling was 99.97%. The results show that the two pipelines exhibit a high degree of consistency, further confirming that ZBOLT strictly follows GATK Best Practices while improving the analysis speed without compromising accuracy. Detailed evaluation results can be found in Table S3.
|
HG001-HG005(ZBOLT vs. GATK) |
SNP | INDEL | ||||
|---|---|---|---|---|---|---|
| Precision | Sensitivity | F-measure | Precision | Sensitivity | F-measure | |
| HG001 | 99.98% | 99.99% | 99.99% | 99.96% | 99.97% | 99.96% |
| HG002 | 99.99% | 99.99% | 99.99% | 99.98% | 99.98% | 99.98% |
| HG003 | 99.99% | 99.99% | 99.99% | 99.97% | 99.96% | 99.97% |
| HG004 | 99.99% | 99.99% | 99.99% | 99.96% | 99.96% | 99.96% |
| HG005 | 99.99% | 99.99% | 99.99% | 99.96% | 99.97% | 99.97% |
3.3 ZBOLT performance in population genomic study
To efficiently analyze the population study consisting of 5,616 high-depth sequencing samples, we deployed the MegaBOLT pipeline on 46 ZBOLT computing servers to run in parallel. With excellent storage density and well-designed layout, the 7.2 Pb storage system, including computing servers and storage devices, was aggregated into only 22U cabinet space, with 16U occupied by the storage device. The significant reduction in cabinet space, compared with conventional methods, alleviates the demands for server room space in big data research.
Data transfer rate is crucial in the big data analysis.33 The ZBOLT system employs multiple 100GB/s transmission channels, optimizing the aggregate bandwidth to 33GB/s. Upon preparing the system, the 1.16Pbp FASTQ files of 5616 WGS samples were transferred to the system for WGS analysis.
The analysis from FASTQ to VCF was successfully completed in 11.6 days, processing over 2.5 Pb of input and output data. On average, the system analyzed 100Tbp of raw sequencing data per day. After the analysis, the energy consumption is measured, with a total consumption of 11,306 kWh and an average consumption of 0.98 kWh per 100Gbp. In other words, the analysis of a WGS sample with 30X sequencing depth consumed approximately 1 kWh.
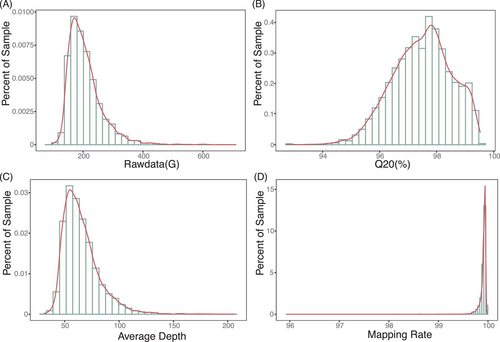
Following the analysis, we calculated the population genomics results. The mean sequencing depth was 68.7X, covering 99.52% of the genome. The mean transition-to-transversion ratio (Ts/Tv) was 1.951 for all SNPs. Representative summary statistics are shown in Figure 4. In total, we called 110 366 736 variants, among which 101 778 160 were SNPs (92.2%), 9 326 668 were Indels, 1 098 959 (0.996%) variants were located in the coding region (Table 4). Overall, the statistic metrics indicated that the population genomics analysis was performed as expected and yielded normal results.
| Consequence | Number of variants |
|---|---|
| Total variants | 110 366 736 |
| SNPs | 101 778 160 |
| Indels | 9 326 668 |
| Coding variation | 1 098 959 |
| Coding sequence variant | 743 |
| Frameshift variant | 25 762 |
| Incomplete terminal codon variant | 26 |
| Inframe deletion | 10 115 |
| Inframe insertion | 4813 |
| Missense variant | 663 556 |
| Protein altering variant | 285 |
| Start lost | 1623 |
| Start retained variant | 23 |
| Stop gained | 17 062 |
| Stop lost | 655 |
| Stop retained variant | 336 |
| Synonymous variant | 373 960 |
4 DISCUSSION
In this study, we explored the ZBOLT analysis pipeline in population genomics studies. Adhering to GATK Best Practices, the ZBOLT analysis system enables a streamlined analysis by running a single command specifying the GATK resource bundle release, eliminating the need for more complicated operations, such as modifying the configure file. In comparison to GATK Best Practices, ZBOLT accelerates the whole analysis process by 10–20 times without sacrificing accuracy.
We assessed the accuracy of ZBOLT using the GIAB benchmark and concordance with GATK Best Practices outputs. Our results demonstrate high accuracy, comparable to that of GATK Best Practices. Although the evaluated results of replicated and controlled experiments showed the same metrics (precision, sensitivity, F-measure), some differences were observed in VCF metrics, these discrepancies were located outside high-confidence region, therefore not affecting the overall evaluation of the pipeline. Furthermore, the concordance evaluation result indicated that ZBOLT is consistent with GATK Best Practices. Overall, our findings suggest that ZBOLT is an accurate and stable analysis system, well-aligned with GATK Best Practices.
In the application of ZBOLT to a newborn genomic study, the system demonstrated its efficiency by analyzing 100Tbp sequencing data per day, highlighting its excellent performance in large-scale population genomic studies. In addition, with a power consumption of 1 kWh per 100Gbp sequencing sample, ZBOLT exhibits advantages in power usage effectiveness,34 making it a more eco-friendly and low-carbon option. The bijou cabinet space required for ZBOLT installation further promotes its widespread application across various population genomics research fields. As an important measure for evaluating big data computing centers, performance, power, area metrics demonstrate that ZBOLT is an exceptional analysis model in the field of population genomics big data research.
Considering the system's throughput and costs, ZBOLT presents significant advantages when applied to population genomic studies comprising more than 5000 WGS, especially for projects involving upwards of 10 000 at 30X coverage.
Currently, the ZBOLT joint calling module is under development and optimization. Upon completion, it will be applied to the joint population analysis. In addition to the analysis pipeline, ZBOLT has developed an integrated system, ZLIMS, which enables automated scheduling of sample extraction, sequencing and analysis. The system can be used in conjunction with a sequencer to tightly integrate sequencing and analysis. Once sequencing is completed, the analysis is automatically initiated, streamlining the research process. Future research directions include innovations in data transmission technology based on compression, which will further accelerate data processing.
Compared with cloud computing solutions, the on-premise ZBOLT analysis system ensures data security and privacy. Moreover, ZBOLT is not reliant on a stable, high-speed network for data transfer, task scheduling and computing, which can be a limiting factor for cloud computing solutions. Users of the ZBOLT system can flexibly control all the software, hardware and data to meet various analysis needs.
To the best of our knowledge, ZBOLT is the first analysis solution specially designed for population genomics study that considers accuracy, efficiency and power consumption. The introduction of ZBOLT is expected to significantly advance population genomics, genetic disease diagnosis and precision medicine.
AUTHOR CONTRIBUTIONS
XJ, MF, BW, and RS conceived and organized this overall population genomic study. MF, YX and ZL designed the detailed research program. YG, XW, GH, and ZL collected the samples and conducted sequencing. YX, SG, JW, WH, LL, JT, QL, XZ, and YZ deployed the ZBOLT system, analyzed the WGS data. ZL, WH, YH carried out the statistical analysis including the evaluation of all outcomes. WZ, LL, XY, and RZ provided relevant computing supports. ZL, XW, and YH wrote the manuscript. MF, YX, and XJ revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
This study was supported by the National Key Research and Development Program of China (grant number: 2022YFC27031020), National Natural Science Foundation of China (grant numbers: 32171441 and 32000398), Natural Science Foundation of Guangdong Province, China (grant number: 2017A030306026), Guangdong-Hong Kong Joint Laboratory on Immunological and Genetic Kidney Diseases (grant number: 2019B121205005), the specific research fund of The Innovation Platform for Academicians of Hainan Province (grant number: YSPTZX202118). This work was supported by China National GeneBank (CNGB). The graphical abstract was created with Biorender.com.
CONFLICT OF INTEREST STATEMENT
The authors declare no potential conflict of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The studies involving human participants were reviewed and approved by the Ethical Clearance the Institutional Review Board of BGI (BGI-IRB 20064). All participants' legal guardians had signed the informed consent form.
Open Research
DATA AVAILABILITY STATEMENT
The HG001-HG005 datasets used for the accuracy evaluation are publicly available and can be accessed and downloaded through GIAB (https://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/). The detailed data path is provided in the Table S1.




