Targeting deubiquitinases for cancer therapy
[Correction added on 30 November 2023; after first online publication missing sub-sections have been added.]
Abstract
The ubiquitin-proteasome system assumes a critical role in numerous cellular processes, and among its components, deubiquitinases (DUBs) have emerged as essential regulators. With roughly 100 DUBs encoded within the human genome, these enzymes can be categorized into two main types: cysteine protease DUBs and metalloproteinase DUBs, based on the catalytic mechanism of the active site. DUBs exert significant influence over specific substrates implicated in cancer progression, establishing them as closely associated with various malignancies, including breast carcinoma, prostate cancer, and chronic myeloid leukemia. Consequently, the targeted inhibition of DUBs presents an enticing therapeutic strategy for cancer treatment. Here, we delve into the functional roles of DUBs in different cancer types and provide a thorough overview of the anticancer properties exhibited by DUB inhibitors. This knowledge will propel the development and clinical application of DUB inhibitors, opening promising avenues for tumor treatment.
1 BACKGROUND
The ubiquitin-proteasome system is a complex machine responsible for the breakdown of 80−90% of abnormally folded or short-lived proteins within cells.1 Ubiquitin, a highly conserved protein consisting of 76 amino acids, undergoes posttranslational modification through the formation of an isopeptide bond between the C-terminus glycine of ubiquitin and the amino acid residues (e.g., lysine) on the substrate. This sequential process involves three enzymes: ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin ligase (E3)2 (Figure 1).
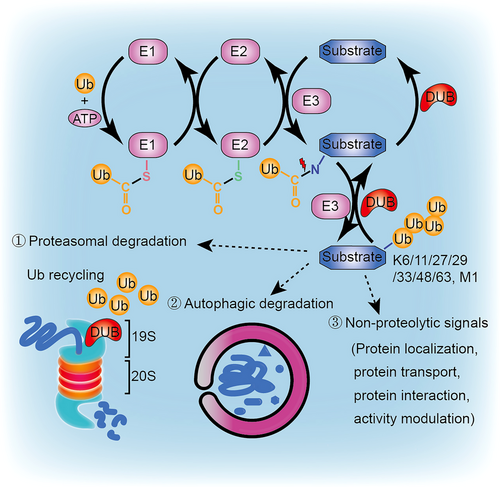
E1 utilizes ATP hydrolysis to establish a thiol ester bond between its active cysteine and the C-terminus of ubiquitin. The activated ubiquitin is subsequently transferred to the active cysteine site in E2, forming a similar thiol ester bond. Finally, E3 confers specificity by recognizing substrates and transferring ubiquitin from E2 to the substrate. Ubiquitin modifications can take the form of mono-ubiquitination, poly-ubiquitination, or heterotypic polyubiquitination, including branched and hybrid chains.3 Through repeated ATP-dependent processes, additional ubiquitins are added to the previously conjugated ubiquitin, generating a polyubiquitin chain. Seven internal lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the N-terminal methionine (M1) allow for diverse chain configurations, with K48 and K63 representing two of the most abundant and well-characterized chain types.3 K48 linkage chains effectively tag substrates for proteasomal degradation, while K63 linkage chains generally serve as signals for autophagic degradation or non-proteolytic functions.4 In addition, atypical ubiquitin modification also occurs on the Cys, Ser, or Thr side chains of targeted proteins.3
Ubiquitination plays an important role in regulating protein localization, transport, interaction, and activity, depending on the specific chain configuration and linkage.5 This process is reversible, as ubiquitin conjugates can be removed from substrates by deubiquitinases/deubiquitinating enzymes (DUBs) (Figure 1). DUBs are vital components of the ubiquitin-proteasome system, responsible for specifically removing ubiquitin from substrates.6 By trimming ubiquitin chains from ubiquitinated proteins, DUBs influence a protein's fate and coordinate proteasomal processing steps. Additionally, DUBs promote ubiquitin recycling by cleaving polymeric ubiquitin chains into ubiquitin monomers. Alterations in DUB activity have been closely associated with cancer, and the presence of DUB inhibitors highlights DUBs as viable and valuable targets for anticancer therapy.
In this review, we will present the recent advancements regarding the role of DUBs in various types of cancer and thoroughly examine the targeted anti-cancer effects demonstrated by DUB inhibitors.
2 CLASSIFICATION AND CATALYTIC ACTIVITY OF DUBS
The human genome encompasses nearly 100 DUBs that exhibit specificity for ubiquitin and can be categorized into nine families: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), JAB1/MPN/Mov34 metalloenzymes (JAMMs), Machado-Josephin domain proteases (MJDs), motif interacting with ubiquitin-containing novel DUB family (MINDYs), ovarian tumor proteases (OTUs), zinc finger-containing ubiquitin peptidase 1 family (ZUP1), monocyte chemotactic protein-induced protein (MCPIP), and after permuted papain fold peptidases of dsRNA viruses and eukaryotes (PPPDE).7-10 The majority of these DUBs belong to the cysteine protease class, with the exception of JAMMs, which are zinc-dependent metalloproteases. These two classes of enzymes exhibit distinct characteristics in terms of ubiquitin-deconjugation (Figure 2). Cysteine protease DUBs possess a nucleophilic cysteine residue in their active site, which facilitates the formation of a covalent intermediate with the substrate. In contrast, metalloprotease DUBs bind to the substrate in a noncovalent manner.
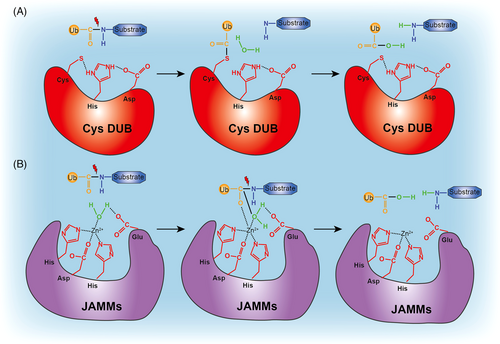
2.1 Cysteine protease DUBs
Most DUBs belong to the cysteine protease class and rely on the thiol group of the active cysteine site for their catalytic activity. Cysteine DUBs utilize a canonical catalytic triad comprising cysteine-histidine-aspartate/asparagine residues to catalyze the hydrolysis of peptide or isopeptide bonds.11 In this process, the histidine residue aids in deprotonating the cysteine, whereas an adjacent aspartate/asparagine residue polarizes the catalytic histidine. During catalysis, the carbonyl carbon atom of the isopeptide peptide bond undergoes nucleophilic attack by the deprotonated cysteine, forming an acyl intermediate transition state. This intermediate, which contains an oxyanion, is stabilized within a region known as the oxyanion hole, which consists of several hydrogen-donating residues. In the subsequent step, the acyl intermediate is hydrolyzed through nucleophilic attack by a water molecule on the carbonyl atom, resulting in the release of free ubiquitin from the enzyme (Figure 2A).
2.2 Metalloprotease DUBs
The JAMMs metalloproteases possess a catalytic core that coordinates Zn2+ and exhibits a catalytic mechanism similar to the metalloprotease thermolysin. Within the catalytic core of JAMMs, Zn2+ is coordinated by two histidine residues, an aspartate residue, and a water molecule.12 Typically, the water molecule is positioned and polarized by an essential glutamate (Glu) residue. In the catalytic process, the Zn2+-bound polarized water molecule performs a nucleophilic attack on the carbonyl carbon atom of the isopeptide bond. This attack results in the formation of a noncovalent intermediate and cleavage of the scissile isopeptide bond (Figure 2B).
3 DUBS IN CANCER
DUBs play a crucial role in various pathological changes, particularly in the context of tumors. The functions of DUBs can vary depending on the type and stage of the tumor. Several DUBs have been categorized as oncogenes or tumor suppressors due to their pivotal roles in tumor development.13 In the following sections, we will provide detailed descriptions of the specific functions of DUBs in both solid tumors, such as breast cancer and prostate cancer, as well as nonsolid tumors, such as chronic myeloid leukemia.
3.1 Breast cancer
Breast cancer stands as the most prevalent malignancy among women globally, characterized by genetic and histopathological variations that enable its classification into five distinct subtypes.14 These subtypes encompass basal-like/triple-negative breast cancer (TNBC), erb-b2 receptor tyrosine kinase 2 (ERBB2/HER2)-enriched, luminal A, luminal B, and normal-like breast cancer. DUBs play a significant role in regulating various pathways implicated in breast cancer, including those involving the estrogen receptor (ER), cell cycle progression, epigenetic modifications, and epithelial–mesenchymal transition (EMT; Figure 3).
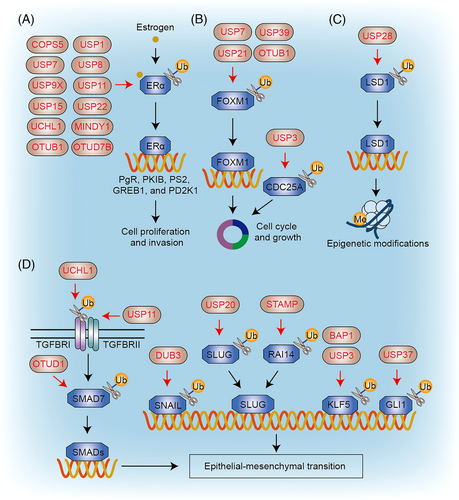
3.1.1 Estrogen receptor
Estrogen receptor alpha (ERα) and beta (ERβ) are pivotal players in the regulation of human breast cancer processes.14 Approximately 70% of diagnosed breast cancers exhibit overexpression of ERα, which acts as an estrogen-activated transcription factor responsible for regulating a vast array of genes.15 Endocrine therapy has shown effectiveness in treating ERα-positive breast cancer; however, the emergence of treatment resistance poses significant challenges in breast cancer treatment. Several mechanisms of resistance have been identified, with dysregulation of ERα signaling being the most common. Hence, understanding the molecular mechanisms underlying ERα signaling is crucial. Notably, numerous DUBs, including COPS5 (CSN5/JAB1), USP1, USP7, USP8, USP9X, USP11, USP15, USP22, UCHL1, MINDY1, OTUB1, and OTUD7B, have been identified as key players in the activation or stabilization of the ERα protein, thereby exerting specific regulatory effects.
USP1, identified as a deubiquitinating enzyme, has been found to regulate the expression of the ERα target gene GREB1 through screening the entire DUB siRNA library in MCF-7 breast cancer cells.16 Notably, USP1 exhibits high expression in breast cancer compared to normal mammary tissues, and its expression correlates with poor patient survival.16 Through its interaction with ERα, USP1 promotes the stability of ERα by removing K48-linked polyubiquitination of the protein. Knockdown of USP1 inhibits ERα signaling, resulting in decreased cell proliferation and invasion in breast cancer cells.16
USP7, USP8, and USP9X can also interact with and deubiquitinate ERα, leading to the stabilization of ERα in MCF-7 and T47D breast cancer cells.17-19 A loss-of-function screen using shRNA targeting all DUBs identifies USP11 as a moderator of ERα transcriptional activity.20 Knockdown of USP11 reduces the expression of ERα target genes, such as PgR, PKIB, and GREB1, as well as cell-cycle-associated genes in LCC1 cells.20 However, the exact underlying mechanism of how USP11 regulates ERα requires further investigation.
Deubiquitinating enzymes USP15 and MINDY1 interact with ERα and remove K48-linked polyubiquitination of the ERα protein, thereby promoting its stabilization in ERα-positive breast cancer cells.21, 22 Similarly, USP22 promotes the stability of the ERα protein by removing both K48- and K63-linked ubiquitination.23 Furthermore, OTUB1 and OTUD7B deubiquitinate and stabilize ERα, leading to increased transcriptional activity of ERα and the expression of target genes, such as PS2, PDZK1, and GREB1, in MCF-7 and T47D breast cancer cells.24, 25 Some deubiquitinating enzymes may also indirectly regulate the levels of ER expression. For instance, UCHL1 decreases ERα levels by mediating the deubiquitination of EGFR, thereby inhibiting ERα transcription.26 Additionally, COPS5 promotes the ubiquitination and proteasomal degradation of NCoR, a key corepressor for ERα.27 In conclusion, these findings suggest that targeting the DUB-ERα complex may represent a potential strategy for treating ERα-positive breast cancer.
3.1.2 Cell cycle
FOXM1 serves as a pivotal transcription factor in driving the growth of basal-like breast cancer, primarily by modulating cell cycle progression. In an RNAi-based screen, USP21 was identified as a DUB that controls the levels of FOXM1 protein.28 USP21 enhances the stability of FOXM1 through binding and deubiquitinating FOXM1.28 Knockdown of USP21 leads to suppression of FOXM1's transcriptional activity, resulting in cell cycle arrest both in vitro and in vivo.28 Additionally, several other DUBs, including USP7, USP39, and OTUB1, are also involved in governing the ubiquitination and degradation of FOXM1.29-31 These findings highlight the significant role of DUBs in regulating the growth induced by FOXM1 activation in basal-like breast cancer.
Furthermore, a genome-level knockout study targeting a set of DUB genes has identified USP3 as a key regulator of CDC25A protein stability.32 Knockdown of USP3 leads to downregulation of CDC25A and a substantial delay in cell cycle progression.32 This, in turn, reduces the growth of cervical tumor xenografts in nude mice.32 The results underscore the crucial role of USP3 in controlling CDC25A stability and its impact on cell cycle progression and tumor growth.
3.1.3 Epigenetic modifications
LSD1 is a lysine-specific histone demethylase that plays a pivotal role in several cellular processes, such as cell stemness, differentiation, and EMT. A siRNA screening has identified USP28 as a bona fide deubiquitinase of LSD1.33 USP28 interacts with and stabilizes LSD1 protein by removing its ubiquitination.33 Knockdown of USP28 results in LSD1 destabilization, leading to the inhibition of cancer stem cell-like characteristics and tumor growth both in vitro and in vivo.33 However, another study has shown that LSD1 is downregulated in breast cancer and suppresses breast cancer metastatic potential.34 These studies reveal a crucial role of USP28 in the epigenetic regulation of breast cancer.
3.1.4 Epithelial–mesenchymal transition
EMT, the process by which epithelial cells obtain mesenchymal characteristics, contributes to various malignant characteristics of cancer cells, including invasion, metastatic potential, and stem-cell-like phenotypes. The progression of breast cancer is driven by the activation of EMT through TGFβ-SMAD signaling.35 TGFβ family members exert their signaling effects through two pairs of transmembrane receptors: type I (TGFBRI) and type II receptor (TGFBRII). Upon activation, TGFBRI propagates the TGFβ signal into the cell, provoking the phosphorylation of receptor-regulated SMADs, such as SMAD2 and SMAD3. Consequently, these SMADs can interact with SMAD4, which then regulate gene expression in the nucleus by recruiting transcription factors, coactivators, and corepressors. Several DUBs, including UCHL1, USP11, and OTUD1, regulate the TGFβ-SMAD signaling pathway in breast cancer.
DUB activity profiling methods have revealed that UCHL1 exhibits pronounced activity in both TNBC cultured cells and tumor tissues in TNBC patients.36 UCHL1 also promotes metastasis in MDA-MB-231 and MDA-MB-436 breast cancer cells in xenograft zebrafish and mice models.36 Functionally, UCHL1 assists TGFβ-SMAD signaling by removing K48-linked polyubiquitination of TGFBRI and SMAD2 proteins.36 Another study reveals that USP11 is associated with EMT in breast cancer based on analysis of published microarray data.37 High USP11 expression correlates with poor survival in human breast cancer patients.37 Moreover, knockdown of USP11 inhibits, whereas overexpression of USP11 promotes cell migration of MDA-MB-231 cells both in vitro and in vivo.37 Mechanistically, USP11 actives SMAD signaling by enhancing the stability of TGFBR2 in SUM159 and MDA-MB-231 breast cancer cells.37 A loss-of-function screen using shRNA targeting 74 DUBs in mice identifies OTUD1 as a new metastasis-repressing factor.38 Low expression of OTUD1 correlates with poor survival in breast cancer patients.38 OTUD1 overexpression increases the expression of the epithelial marker (e.g., CDH1), whereas concurrently decreasing the expression of the mesenchymal markers (e.g., CDH2).38 Mechanically, OTUD1 deubiquitinates SMAD7, an inhibitor of the TGFβ pathway, leading to enhanced stabilization of SMAD7.38
Several other DUBs, including DUB3, USP20, BAP1, USP3, and STAMBP, have been reported to regulate EMT in breast cancer by targeting EMT-related transcription factors, such as SNAIL1, SLUG, and KLF5. The cytokine IL-6 induces the expression of DUB3, which, in turn, deubiquitinates and stabilizes SNAIL1 in breast cancer.39 In addition, CDK4/6 phosphorylates DUB3 at Ser41, activating DUB3 and promoting SNAIL1 protein level.40 Gain- and loss-of-function screens have identified USP20 as a key DUB of SLUG in breast cancer.41 USP20 stabilizes SLUG, resulting in cell migration, invasion, and metastasis of breast cancer.41 Higher USP20 expression is associated with poor prognosis of breast cancer patients.41 A siRNA library screen identifies BAP1 and USP3 as bona fide DUBs of KLF5.42, 43 BAP1 and USP3 stabilize KLF5 by removing K48-linked polyubiquitin chains from KLF5 in breast cancer.42, 43 STAMBP, a member of the DUB family characterized by its JAMM metalloproteinase activity, promotes human TNBC tumor growth by inhibiting the K48-linked ubiquitination of RAI14, thereby preventing its degradation and stabilizing RAI14 protein.44 The depletion of STAMBP induces the downregulation of EMT-related transcription factors such as SLUG, lead to inhibition of TNBC tumor growth.44 These findings indicate that DUBs could serve as potential therapeutic targets for interfering with EMT in breast cancer.
The Hedgehog signaling pathway also plays a role in regulating EMT during breast cancer development.45 In the canonical pathway, Hedgehog ligands bind to the receptor PTCH, leading to the activation of downstream signaling mediated by SMO. The activation of SMO triggers the nuclear translocation of glioma-associated oncogene homolog (GLI) transcription factor, which regulates the expression of target genes, including c-MYC, BCL2, and SNAIL1. USP37 interacts with and stabilizes GLI1 in breast cancer.46 Accordingly, USP37 promotes EMT and stemness in breast cancer cells.46 Conversely, knockdown of USP37 decreases GLI1 and inhibits tumorigenicity of breast cancer in vivo.46 These findings indicate that blocking the USP37-Hedgehog signaling could inhibit EMT in breast cancer.
3.2 Prostate cancer
Prostate cancer ranks as the second most prevalent cancer among men worldwide and is responsible for being the fifth leading cause of death globally.47 The risk of developing prostate cancer tends to increase with age.47 DUBs play a significant role in modulating multiple pathways associated with prostate cancer, including those involving the androgen receptor (AR), epigenetic modifications, and chemotherapy resistance (Figure 4).
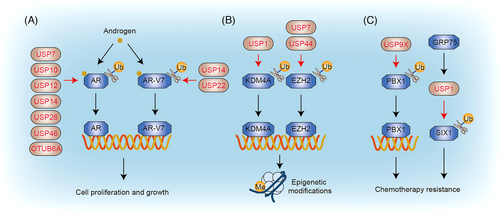
3.2.1 Androgen receptor
AR is a steroid receptor transcription factor that is stimulated by androgenic hormone ligands, including testosterone and dihydrotestosterone. Upon binding with its ligand, AR undergoes translocation from the cytoplasm to the nucleus and attaches to the androgen-response element within the promoter regions of target genes. AR plays a crucial role in regulating gene expression with diverse functions that promote the development of prostate cancer.48 Androgen deprivation therapy, aimed at suppressing the AR signaling axis, is commonly used as a first-line treatment for prostate cancer. However, resistance to these therapies can develop in some patients, partly due to the presence of constitutively active spliced AR variants, such as AR-V7. Emerging evidence suggests that specific DUBs regulate AR protein stability and activity. Several DUBs, including USP7,49 USP12,50 USP14,51 USP26,52 USP46,53 and OTUD6A,54 are essential regulators of AR stabilization in prostate cancer. Additionally, USP10 associates with the AR and stimulates its transcriptional activity,55 although its impact on the ubiquitylation status of AR remains unclear. Furthermore, two deubiquitinases, USP14 and USP22, have been found to promote the stability of AR-V7 through their deubiquitinase activities.56
3.2.2 Epigenetic modifications
The overexpression of lysine-specific demethylase 4A (KDM4A), a histone demethylase, plays a significant role in various cancers. Studies have reported the upregulation of KDM4A expression in prostate tissues of phosphatase and tensin homolog (PTEN) knockout mice.57 Deletion of KDM4A inhibits the proliferation and survival of prostate cancer cells in both in vivo and in vitro experiments.57 The deubiquitinating enzyme USP1 manipulates the K48-linked ubiquitination and stability of KDM4A.57 Increased expression of KDM4A and high levels of USP1 are observed in most prostate tumors.57 Inhibiting USP1 promotes the response of prostate cancer cells to the therapeutic agent enzalutamide.57
USP7 is involved in the progression and metastasis of various malignant tumors by regulating the stability of target proteins. For instance, USP7 targets the enhancer of zeste homolog 2 (EZH2), an enzyme that catalyzes the methylation of the lysine 27 site of histone H3, and stabilizes it through deubiquitination.58 In prostate cancer cells, knockdown of USP7 inhibits the transcriptional inhibition of EZH2.58 Moreover, ectopic introduction of EZH2 restores cell migration, invasion, and proliferation in prostate cancer cells, while USP7 knockdown reduces the progression of prostate cancer.58 USP44-mediated deubiquitination also upregulates the protein stability of EZH2.59 Knockdown of USP44 inhibits the tumorigenic properties and cancer stem-like behavior of prostate cancer cells.59 The inhibition of tumorigenesis induced by USP44 knockdown can be restored by the ectopic introduction of EZH2.59
3.2.3 Chemotherapy resistance
Chemotherapy resistance poses a significant challenge to the effective treatment of advanced prostate cancer. Targeting DUBs holds promise in overcoming chemotherapy resistance in prostate cancer. The transcription factor PBX1 promotes the proliferation of prostate cancer cells and confers resistance to common anticancer drugs such as doxorubicin and cisplatin.60 The stability of PBX1 is regulated by the ubiquitin-proteasome pathway, wherein the deubiquitinating enzyme USP9X interacts with PBX1 protein and stabilizes it by reducing K48-linked polyubiquitination.60 In addition, USP9X inhibitor significantly induced PBX1 degradation and promoted apoptosis of chemoresistant prostate cancer cells.60
In some cases, patients diagnosed with advanced castration-resistant prostate cancer may develop an independent phenotype that is not dependent on the androgen receptor (AR). The chaperone GRP75 recruits USP1 to inhibit K48-linked polyubiquitination of SIX1, thereby promoting castration resistance in prostate cancer.61 In vitro and xenograft mouse models demonstrate that genetic or pharmacological inhibition of USP1 functionally inhibits tumor growth and combats castration resistance in prostate cancer cells.61
3.3 Chronic myeloid leukemia
Chronic myeloid leukemia (CML) is a malignant clonal disease characterized by the abnormal growth of hematopoietic stem cells.62 It leads to an increase in myeloid cells, red blood cells, and platelets in peripheral blood, along with significant myeloid proliferation in the bone marrow.62 The Bcr-Abl fusion protein, characterized by its constitutively activated kinase activity, plays a critical role in the transformation of hematopoietic cells, making it an attractive therapeutic target for CML. Within the context of CML, DUBs exert regulatory control over crucial players such as Bcr-Abl (Figure 5).

USP7 interacts with Bcr-Abl, blocking its polyubiquitination and degradation in CML cells.63 Chemical or genetic inhibition of USP7 can trigger the breakdown of Bcr-Abl protein, inhibit Bcr-Abl function, and induce apoptosis of CML cells.63 Additionally, the antimalarial agent artesunate inhibits the interaction between USP7 and BCR-ABL, thereby reducing stability of Bcr-Abl protein and inducing apoptosis in CML.63 Moreover, the USP9X inhibitor WP1130 induces modifications in Bcr-Abl protein through K63-linked ubiquitin polymers, causing Bcr-Abl to aggregate and be unable to transmit signals, ultimately leading to apoptosis.64
Other studies have revealed that caspase activation, either through proteasome inhibition or DUBs inhibition, downregulates Bcr-Abl expression by inducing Bcr-Abl cleavage.65 S-phase kinase-associated protein 2 (SKP2) is one of the E3 ligases that serves as a regulator of Bcr-Abl by facilitating its K63-linked ubiquitination and subsequent activation.66 Furthermore, USP10, functioning as a SKP2 deubiquitinase, amplifies Bcr-Abl activation by mediating the deubiquitination and stability of SKP2 in CML cells.66 Inhibition of USP10 profoundly restrains the growth of both imatinib-sensitive and imatinib-resistant CML cells.66
4 THERAPEUTIC POTENTIAL OF DUB INHIBITORS
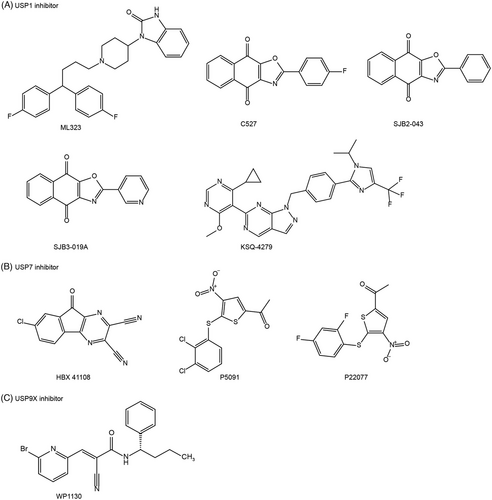
DUBs play significant roles in regulating the activity and stabilization of specific substrates, which are involved in various cellular processes associated with cancer, such as cell-cycle control, proliferation, differentiation, and DNA damage response. In this context, we will primarily focus on exploring the therapeutic potential of different DUB inhibitors for cancer treatment. These inhibitors can be categorized into proteasomal DUB inhibitors (such as Rpn11, USP14, and UCHL5) (Figure 6) and nonproteasomal DUB inhibitors (including USP1, USP7, and USP9X; Figure 7). Rpn11 belongs to JAMM metalloprotease DUBs, whereas USPs and UCHs are categorized as cysteine protease DUBs. By targeting specific DUBs, these inhibitors hold promise as potential treatments for cancer.
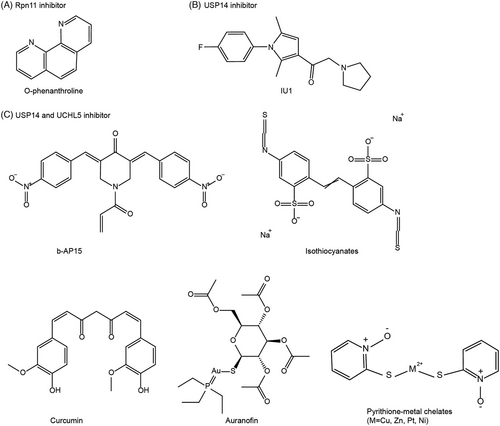
4.1 Inhibitors of proteasomal DUBs
The 26S proteasome consists of a proteolytic core called the 20S proteasome and one or two regulatory caps known as the 19S proteasome. Within the 19S proteasome, three DUBs are associated: Rpn11/POH1/PSMD14, USP14/Ubp6, and UCHL5/UCH37. Rpn11 is responsible for removing the entire poly-Ub chain at the base of the substrate, while USP14 and UCHL5 edit the poly-Ub chains by cleaving them from the distal end.67
4.1.1 Rpn11 inhibitor
Rpn11 possesses DUB isopeptidase activity, which is mediated by a zinc-binding JAMM domain. Rpn11 is elevated in various cancer types, such as ovarian carcinoma, leukemia, melanoma cells, and multiple myeloma, with higher expression of Rpn11 correlating with poor patient survival.68 O-phenanthroline is a heterocyclic organic compound that chelates the Zn2+ required for the catalytic activity of Rpn11 (Figure 6A). Pharmacological inhibition of Rpn11 with o-phenanthroline has been shown to induce apoptosis and combat resistance to bortezomib in multiple myeloma cells.69 Accordingly, o-phenanthroline triggers an induction of tumor suppressor proteins, including p53 and hDLG1.69 Given the effectiveness of proteasome inhibitors in cancer therapy, Rpn11 inhibitors hold promise as potential candidates. However, the efficacy and toxicity of Rpn11 inhibitors in cancer therapy are yet to be determined.
4.1.2 USP14 inhibitor
USP14 has been implicated in promoting tumorigenesis in various cancer types, including prostate cancer cells, breast cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma, and gastric cancer.70-72 Through its DUB activity, USP14 can interact with and stabilize AR proteins.73, 74 IU1 stands as a specific inhibitor targeting the activity of USP14 (Figure 6B). Mechanically, the benzene ring of IU1 forms interactions with His426, Tyr436, and Tyr476 in USP14.75 Inhibition of USP14 using IU1 has been shown to accelerate the degradation of AR, leading to AR-dependent cell cycle arrest in prostate cancer cells and AR-positive breast cancer cells.73, 74 IU1 has also demonstrated the ability to suppress proliferation and increase IDO1 ubiquitination, thereby enhancing colorectal cancer sensitivity to anti-PD-1 therapy.76 These findings suggest that targeting USP14 holds promise as a therapeutic approach in cancer treatment.
4.1.3 Dual inhibitors of USP14 and UCHL5
A specific inhibitor of USP14 and UCHL5, b-AP15, was identified in a screening for agents inducing cathepsin D-dependent apoptosis77 (Figure 6C). b-AP15 possesses an α, β-unsaturated carbonyl moiety, which is highly prone to undergoing a Michael addition reaction with nucleophilic cysteine thiolates of DUBs. b-AP15 has demonstrated the ability to induce strong oxidative stress and cell death in various cancer cells and inhibit tumor growth in mice.78-80 Preclinical studies have also shown that USP14 and UCHL5 inhibitors exhibit broad antitumor activity against both nonsolid and solid tumors (e.g., multiple myeloma and hypopharyngeal carcinoma), overcoming resistance to bortezomib.78, 81 This indicates that their mechanism of action against cancer is distinct from that of 20S proteasome inhibitors.78, 81 Notably, VLX1570, a derivative of b-AP15, underwent investigation in a Phase I clinical trial for relapsed multiple myeloma.82 However, the trial was halted prematurely due to the serious side effects.82
Some natural products (e.g., curcumin and isothiocyanates) have been reported to inhibit USP14 and UCHL5 (Figure 6C). Curcumin and its analog AC17 have been demonstrated to impair the USP14 and UCHL5 activity through its α,β-unsaturated ketone structure, resulting in the accumulation of ubiquitinated proteins in lung cancer cells.83 Isothiocyanates, abundant in cruciferous vegetables, are natural products containing the chemical group –N = C = S. Isothiocyanates have been reported to inhibit the growth of breast and pancreatic cancer both in vitro and in vivo.84
Furthermore, various metal complexes, including auranofin and pyrithione-metal chelates, have been identified as potent inhibitors of USP14 and UCHL5 (Figure 6C). Auranofin, a clinically used gold (I)-containing anti-rheumatoid arthritis drug, exhibits promising anticancer activities. Apart from targeting thioredoxin reductase (TrxR), auranofin has been evaluated as a potential inhibitor of the proteasomal DUBs USP14 and UCHL5, effectively inducing liver and breast cancer cell death.85 Au atoms present in auranofin demonstrate an affinity for binding to the active sites of both USP14 and UCHL5 enzymes.85 Consequently, auranofin enhances the protein levels of proteasome substrates, including p21 and IKBα.85 Additionally, pyrithione-metal chelates, such as copper, zinc, platinum, and nickel, can target USP14 and UCHL5, resulting in the accumulation of ubiquitinated proteins and cytotoxic effects in various cancer cells, including liver cancer, lung cancer, multiple myeloma, and leukemia.86-89 Therefore, proteasomal DUB inhibitors hold potential as new therapeutic options for cancer.
4.2 Inhibitors of nonproteasomal DUBs
4.2.1 USP1 inhibitor
When USP1 forms a heterodimeric complex with USP1-associated factor 1 (UAF1), the DUB activity of USP1 is significantly enhanced.90 This USP1/UAF1 complex plays a critical role in maintaining genomic integrity by removing ubiquitin from FA complementation group D2 (FANCD2) and the proliferating cell nuclear antigen (PCNA).91, 92 Pimozide, a medication clinically used for Tourette syndrome, inhibits the DUB activity of USP1, resulting in increased levels of PCNA and FANCD2 mono-ubiquitination.93 Acting as an USP1 inhibitor, pimozide, a diphenyl-butylpiperidine compound, effectively overcomes cancer cell resistance to cisplatin and demonstrates potent inhibition of lung tumor growth both in vitro and in vivo.94 Similarly, other USP1 inhibitors, including ML323, C527, SJB2-043, SJB3-019A, and KSQ-4279, have shown the ability to induce cell cycle arrest and cell death in a spectrum of malignancies, encompassing esophageal squamous cell carcinoma, leukemia, and multiple myeloma, both in vitro and in vivo settings95-98 (Figure 7A). Hence, targeting USP1 presents a promising opportunity for anti-cancer therapy.
4.2.2 USP7 inhibitor
USP7 (HAUSP) plays a crucial role in deubiquitinating p53and targeting USP7 with inhibitors may hold promising potential for antitumor activity by stabilizing p53.99 Several USP7 inhibitors, such as HBX 41108, P5091, and P22077, have demonstrated the ability to stabilize p53 and activate the transcription of p53 target genes (e.g., p21 and bax) in colon carcinoma, multiple myeloma, and neuroblastoma cells100-102 (Figure 7B). As a cyano-indenopyrazine derivative, HBX 41108 preferentially interacts with a small hydrophobic pocket in the USP7-ubiquitin complex, leading to the inhibition of USP7 activity.100 These findings highlight the therapeutic potential of USP7 inhibitors in promoting p53-mediated tumor suppression.
Furthermore, USP7 inhibitors have also shown the capability to promote the degradation of PD-L1 in gastric cancer cells, leading to enhanced antitumor immune responses.103 This involvement of USP7 in cancer immunotherapy further supports the notion that USP7 inhibitors could be of great benefit in the treatment of cancer.
4.2.3 USP9X inhibitor
USP9X exhibits dual roles as both an oncoprotein and a tumor suppressor, depending on the specific tumor types and cellular context. USP9X functions as a tumor-promoting factor in numerous human malignancies, such as colon cancer, breast cancer, and lymphomas.104-106 The first-described USP9X inhibitor, WP1130 (Degrasyn), also inhibits other DUBs, including USP5, USP14, and UCHL5107 (Figure 7C). WP1130 exerts its effects by covalently modifying the active cysteine residues of DUBs, owing to the presence of the α, β-unsaturated carbonyl group. By inhibiting USP9X, WP1130 suppresses the deubiquitination of several kinases, including Bcr-Abl and Jak2, leading to their translocation to the aggresome and subsequent blocking of their signal transduction activities in multiple myeloma and CML cells.64, 108 Additionally, USP9X removes polyubiquitin chains from the antiapoptotic proteins MCL1 and XIAP, preventing their proteasomal degradation.105, 109 Therefore, inhibition of USP9X by WP1130 sensitizes B-cell lymphoma cells to anti-cancer agents.105, 109
However, despite the aforementioned findings pointing towards the oncoprotein roles of USP9X, several studies have revealed that USP9X actually suppresses tumorigenesis of pancreatic cancer by deubiquitinating substrates such as LATS kinases.110 These contrasting findings highlight the complex and context-dependent role of USP9X in cancer.
5 CONCLUSION
DUBs can regulate diverse pathways in various cancer types, playing crucial roles in tumor cell cycle, cell death, EMT, DNA repair, and other processes. In this review, we have highlighted the different regulatory pathways of DUBs in breast carcinoma, prostate cancer, and chronic myeloid leukemia, as well as the proposed anti-tumor strategies targeting these DUBs. While encouraging antitumor effects have been observed with DUB inhibitors, several challenges need to be addressed in the pursuit of targeting DUBs for cancer therapy.
One major challenge is the exploration of substrate specificity among DUBs. With a smaller quantity of DUBs (∼100) compared with E3 ligases (∼700), a single DUB often regulates the ubiquitination of multiple substrates. Furthermore, different DUBs are often involved in the regulation of the same substrate, adding complexity to the temporal and spatial removal of ubiquitin modifications. Understanding how these DUBs work together in a coordinated manner remains an intriguing question, and further studies is essential to investigate their combined effects on the same protein under unified conditions.
Additionally, there are still DUBs that lack specific targeted inhibitors or whose antitumor mechanisms have not been fully elucidated. High-throughput screening and structure optimization can contribute to the discovery of novel DUB inhibitors, whereas selective DUB inhibitors will help unravel the distinct roles of DUBs within the regulatory network. Further research and exploration are necessary to expand our knowledge of DUBs and uncover their full potential in cancer therapeutics.
In conclusion, the field of DUBs holds great promise for cancer treatment due to their involvement in critical cellular processes. As we continue to unravel the complexities of DUB-mediated regulation in cancer, it is crucial to address the challenges ahead, deepen our understanding of substrate specificity, discover more specific inhibitors, and elucidate the precise roles of individual DUBs. These endeavors will pave the way for the advancement of targeted therapies and ultimately improve cancer patient outcomes in the future.
AUTHOR CONTRIBUTIONS
X.C. and J.L. contributed to the conception of the review. Q.X. contributed to the manuscript preparation. X.C. and D.T. edited the manuscript. All authors have read and approved the review.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81972399 and 82272660), the Natural Science Foundation Research Team of Guangdong Province (2018B030312001), the Plan on Enhancing Scientific Research in GMU (02-410-2302289XM), the Basic and Applied Basic Research Project of the Guangzhou Basic Research Program (202201011411), and Guangdong Basic and Applied Basic Research Foundation (2021A1515011382).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS APPROVAL
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.




