Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma
Abstract
Background
Resistance to ferroptosis, a regulated cell death caused by iron-dependent excessive accumulation of lipid peroxides, has recently been linked to lung adenocarcinoma (LUAD). Intracellular antioxidant systems are required for protection against ferroptosis. The purpose of the present study was to investigate whether and how extracellular system desensitizes LUAD cells to ferroptosis.
Methods
Established human lung fibroblasts MRC-5, WI38, and human LUAD H1650, PC9, H1975, H358, A549, and H1299 cell lines, tumor and matched normal adjacent tissues of LUAD, and plasma from healthy individuals and LUAD patients were used in this study. Immunohistochemistry and immunoblotting were used to analyze protein expression, and quantitative reverse transcription-PCR was used to analyze mRNA expression. Cell viability, cell death, and the lipid reactive oxygen species generation were measured to evaluate the responses to ferroptosis. Exosomes were observed using transmission electron microscope. The localization of arachidonic acid (AA) was detected using click chemistry labeling followed by confocal microscopy. Interactions between RNAs and proteins were detected using RNA pull-down, RNA immunoprecipitation and photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation methods. Proteomic analysis was used to investigate RNA-regulated proteins, and metabolomic analysis was performed to analyze metabolites. Cell-derived xenograft, patient-derived xenograft, cell-implanted intrapulmonary LUAD mouse models and plasma/tissue specimens from LUAD patients were used to validate the molecular mechanism.
Results
Plasma exosome from LUAD patients specifically reduced lipid peroxidation and desensitized LUAD cells to ferroptosis. A potential explanation is that exosomal circRNA_101093 (cir93) maintained an elevation in intracellular cir93 in LUAD to modulate AA, a poly-unsaturated fatty acid critical for ferroptosis-associated increased peroxidation in the plasma membrane. Mechanistically, cir93 interacted with and increased fatty acid-binding protein 3 (FABP3), which transported AA and facilitated its reaction with taurine. Thus, global AA was reduced, whereas N-arachidonoyl taurine (NAT, the product of AA and taurine) was induced. Notably, the role of NAT in suppressing AA incorporation into the plasma membrane was also revealed. In pre-clinical in vivo models, reducing exosome improved ferroptosis-based treatment.
Conclusion
Exosome and cir93 are essential for desensitizing LUAD cells to ferroptosis, and blocking exosome may be helpful for future LUAD treatment.
Abbreviations
-
- AA
-
- arachidonic acid
-
- AAV5
-
- adeno-associated virus 5
-
- ACSL4
-
- acyl-CoA synthetase long-chain family member
-
- AdA
-
- adrenal acid
-
- ALIX
-
- ALG-2 interacting protein X
-
- ALK
-
- anaplastic lymphoma kinase
-
- ANOVA
-
- analysis of variance
-
- AP-2α
-
- transcription factor AP-2 alpha
-
- BME
-
- basement membrane extract
-
- BSA
-
- bovine serum albumin
-
- CAFs
-
- cancer-associated fibroblasts
-
- CD63
-
- cluster of differentiation 63
-
- CD9
-
- cluster of differentiation 9
-
- CDO1
-
- cysteine dioxygenase type 1
-
- CDR1as
-
- circRNA cerebellar degeneration-related protein 1 transcript antisense
-
- CDX
-
- cell-derived xenograft
-
- ceRNAs
-
- competing endogenous RNAs
-
- cir34
-
- circRNA_100934
-
- cir93
-
- circRNA_101093
-
- circRNA
-
- circular RNA
-
- CSAD
-
- cysteine sulfinic acid decarboxylase
-
- DBD
-
- DNA binding domain
-
- DFO
-
- deferoxamine
-
- DIG
-
- digoxigenin
-
- DMA
-
- 5,5-(N-N-dimethyl)-amiloride hydrochloride
-
- DMEM
-
- Dulbecco's modified eagle medium
-
- DMSO
-
- dimethyl sulfoxide
-
- EGFR
-
- epidermal growth factor receptor
-
- ELISA
-
- enzyme linked immunosorbent assay
-
- EpCAM
-
- epithelial cell adhesion molecule
-
- ESI
-
- electrospray ionization
-
- F
-
- phenylalanine
-
- FABP3
-
- fatty acid-binding protein 3
-
- FABPs
-
- fatty acid-binding proteins
-
- FBS
-
- fetal bovine serum
-
- FDA
-
- Food and Drug Administration
-
- Fer-1
-
- Ferrostatin-1
-
- FISH
-
- fluorescence in situ hybridization
-
- GC-MS
-
- gas chromatography-mass spectrometry
-
- GFP
-
- green fluorescent protein
-
- HAND1
-
- heart and neural crest derivatives expressed 1
-
- HIC1
-
- hypermethylated in cancer 1
-
- HNF4a
-
- hepatocyte nuclear factor alpha
-
- HPLC
-
- high performance liquid chromatograph
-
- HRP
-
- horseradish peroxidase
-
- IB
-
- immunoblotting
-
- IF
-
- immunofluorescence
-
- IKZF1
-
- IKAROS family zinc finger 1
-
- IKZF2
-
- IKAROS family zinc finger 2
-
- IHC
-
- immunohistochemistry
-
- kRAS
-
- v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
-
- Lipro-1
-
- liproxstatin-1
-
- LPCAT3
-
- lysophosphatidylcholine acyltransferase 3
-
- LUAD
-
- lung adenocarcinoma
-
- m6A
-
- N6-methyladenosine
-
- MDA
-
- malondialdehyde
-
- MDM2
-
- mouse double minute 2 homolog
-
- MS
-
- mass spectrometry
-
- MSCs
-
- mesenchymal stem cells
-
- NAT
-
- N-arachidonoyl taurine
-
- NUP107
-
- nucleoporin 107
-
- ORF
-
- open reading frame
-
- OS
-
- overall survival
-
- p53
-
- tumor protein 53
-
- PAR-CLIP
-
- photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation
-
- PBS
-
- phosphate-buffered saline
-
- PDX
-
- patient-derived xenograft
-
- PFA
-
- paraformaldehyde
-
- PI
-
- propidium iodide
-
- PKH67
-
- Paul Karl Horan-67
-
- PKE
-
- piperazine erastin
-
- PLTP
-
- phospholipid transfer protein
-
- PUFA
-
- poly-unsaturated fatty acids
-
- qPCR
-
- quantitative RT-PCR
-
- RIP
-
- RNA, immunoprecipitation
-
- ROS
-
- reactive oxygen species
-
- RSL3
-
- ras-selective lethal small molecule 3
-
- S
-
- serine
-
- SDS
-
- sodium dodecyl sulfate
-
- SSC
-
- saline sodium citrate
-
- TCEP
-
- tris(2-chloroethyl) phosphate
-
- TEM
-
- transmission electron microscope
-
- TLX2
-
- T cell leukemia homeobox 2
-
- TMB
-
- 3,3′,5,5′-tetramethylbenzidine
-
- TRPV1
-
- transient receptor potential vanilloid 1
-
- TSG101
-
- tumor susceptibility 101
-
- UV
-
- ultraviolet
-
- αSMA
-
- alpha smooth muscle actin
-
- 4-HNE
-
- anti-4-hydroxynonenal
-
- 4-SU
-
- 4-thiouridine
1 BACKGROUND
Lung cancer is a leading cause of cancer death worldwide, with lung adenocarcinoma (LUAD) being the most prevalent subtype [1, 2]. Although driver mutations, such as those in epidermal growth factor receptor (EGFR), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (kRAS), anaplastic lymphoma kinase (ALK), and tumor protein 53 (TP53), are major causes of LUAD [3-5], other mechanisms, including the dysregulation of N6-methyladenosine (m6A) RNA methylation, are critical for LUAD tumorigenesis [6, 7]. Recently, resistance to ferroptosis, a newly identified type of regulated cell death caused by iron-dependent excessive accumulation of lipid peroxides, has been regarded as another non-mutation mechanism linked to the initiation and progression of LUAD [7-9]. Although enhanced cell metabolic activity enables tumor cells to produce more lipid peroxides [10], the existence of intracellular anti-oxidant system strongly protects LUAD cells against ferroptosis [6, 7, 9]. However, prior studies have mainly focused on the effects of intracellular antioxidant systems, whether extracellular systems also desensitize LUAD cells to ferroptosis and the underlying mechanisms remain unclear.
Exosome is a type of extracellular vesicle that originated from endocytosis [11]. As a master regulator of cellular signaling, exosome orchestrates various autocrine and paracrine functions to alter tumor microenvironments, growth and progression [12]. Because exosome is taken by target cells and secreted from host cells, they are critical messengers that participate in cell-to-cell communications [13]. Interestingly, one study revealed that ferroptosis resistance in tumor cells was achieved through the formation of ferritin-containing exosomes that removed labile iron from the cell [14]. However, the effects of extracellular exosome uptake on the sensitivity of LUAD cells to ferroptosis remain unexplained.
In addition to metabolites, such as iron, circular RNAs (circRNAs) are important components in exosome [15]. Increasing studies have demonstrated that circRNAs are closely linked to LUAD tumorigenesis [16-18]. circRNAs are more stable than other types of RNAs because of their covalently closed loop structure [19]. The protection from vesicles further increases the stability of circRNAs and enhances their efficiency in transmitting information among cells [20]. A few studies have demonstrated that exosome suppresses ferroptosis in miRNA- or protein-dependent manners [21, 22]. However, our understanding of the roles of circRNAs, especially exosomal circRNAs in modulating the sensitivity of LUAD cells to ferroptosis remains quite limited.
Although the functions of circRNAs as competing endogenous RNAs (ceRNAs) are well established, the interactions between circRNAs and proteins are less understood [23]. Ferroptosis is closely associated with metabolism [24]; therefore, proteins that both interact with circRNAs and participate in metabolism provide promising candidates to influence ferroptosis. Fatty acid-binding proteins (FABPs) are critical for the transport of poly-unsaturated fatty acids (PUFA) to specific cellular compartments [25]. Notably, the peroxidation of PUFAs in the plasma membrane, mainly arachidonic acid (AA) and adrenal acid (AdA), is essential for sensitizing cells to ferroptosis [26]. However, whether and how circRNA-FABP interactions regulate PUFAs and ferroptosis sensitivity remain largely unknown. Therefore, we set up to investigate whether and how exosome and circRNA desensitize LUAD cells to ferroptosis.
2 MATERIALS AND METHODS
2.1 Cell culture
Established human lung fibroblast MRC-5, WI38, and human LUAD H1650, PC9, H1975, H358, A549, and H1299 cell lines were purchased from Fuheng Biotechnology (Shanghai, China) and validated by short tandem repeat analysis. For monolayer culture, cells were cultured with Dulbecco's modified eagle medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Sage Creation Science Co. Ltd, Beijing, China) and 1% penicillin/streptomycin (Hyclone).
Patient-derived primary LUAD cells were derived from LUAD tissues (collected at Department of Bio-bank, Shanghai Chest Hospital, Shanghai, China at March 2020). Briefly, fresh LUAD tissues without necrosis were washed with ice-cold Dulbecco's phosphate buffered saline (Sigma, St Louis, MO, USA) for three times before being re-suspended in DMEM containing collagenase I (2 mg/mL, Solarbio, Shanghai, China) at 37°C in 5% CO2 for 4 h. After an additional three washes with fresh DMEM, cells were cultured in routine conditions for 1 week. Subsequently, tumor epithelial cells were sorted using anti-epithelial cell adhesion molecule (EpCAM) antibodies (#FAB9601G; R&D Systems, Minneapolis, MN, USA) by a flow cytometer to obtain EpCAM (+) cells, and the remaining cells were considered EpCAM (-) cells. For 3-dimentional (3D) spheroid culture, basement membrane extract (BME; Trevigen, Gaithersburg, MD, USA) was seeded in a 96-well plate at 50 μL/well and incubated at 37°C for 30 min. Then, cells were seeded onto the BME at a density of 1 × 105 cells per well. After the formation of spheroids, they were treated with dimethyl sulfoxide (DMSO; Beyotime, Haimen, Jiangsu, China), erastin (Sigma) or ras-selective lethal small molecule 3 (RSL3, Sigma). Images were captured after staining with SYTOXTM green (Invitrogen, Carlsbad, CA, USA).
2.2 Animals
All athymic nude mice (6-week-old) were purchased from Jiesijie (Shanghai, China). They were bred and housed in specific pathogen-free (SPF) animal facilities. All animal experimental procedures were conducted according to the animal welfare guidelines and based on the “3R” principle (reduction, replacement, refinement) and approved by the Animal Care Committee of Shanghai Chest Hospital.
To generate routine cell-derived xenograft (CDX) mouse models, established LUAD cells (initial 5 × 106) were subcutaneously injected into the bilateral dorsal flank of athymic nude mice. The mice were euthanized by cervical dislocation at 18, 36 or 80 days after injection.
To generate H1975/A549 cell-implanted intrapulmonary LUAD mice, athymic nude mice were intrapulmonarily injected with cells (5 × 106) under anesthesia and then intranasally administered adeno-associated virus 5 (AAV5) particles (2 × 1012 viral particles/mL, Genomeditech, Shanghai, China) 3 weeks later. The mice were euthanized at 36 days after injection.
To generate patient-derived xenograft (PDX) mouse models, fresh LUAD tissues with a size of 2-3 mm3 were subcutaneously implanted into athymic nude mice. After successful passage, the PDX mice were used for further studies. Tumor volumes were calculated as 0.5 × L × W2 (L indicating length while W indicating width). For drug administration, piperazine ketone erastin (PKE, MedChemExpress, Monmouth Junction, NJ, USA) and GW4869 (Sigma) were injected. The mice were euthanized at 32 or 80 days after passage.
2.3 Patient specimens and information
All tissue specimens were collected at Department of Bio-bank, Shanghai Chest Hospital between May 2013 and December 2020. All plasma specimens were collected at Department of Bio-bank and Department of Laboratory Medicine, Shanghai Chest Hospital between July 2014 and December 2020. Tumor sections were reviewed by experienced pathologists at Shanghai Chest Hospital according to the World Health Organization classification criteria. Tissues were formalin-fixed paraffin-embedded, or made to ex vivo slice as previously described [27], or homogenized and lysed using appropriate lysis buffer. Mutations in tissues were sequenced by Mapbioo Biotechnology (Shanghai, China). Informed written consent forms were obtained from all patients. All protocols for human studies were approved by the institutional ethics committee of Shanghai Chest Hospital.
Tissues from LUAD patients (n = 250) were used for evaluating malondialdehyde (MDA) and circRNA_101093 (cir93), and correlations among taurine, FABP3 and intracellular cir93 level. The patient information is summarized in Supplementary Table S1.
Tissues from LUAD patients (n = 188) were used for analyzing 4-hydroxynonenal (4-HNE) by immunohistochemistry (IHC). The patient information is summarized in Supplementary Table S2.
Plasma exosomes from healthy individuals (n = 200) and LUAD patients (n = 200) were used for measuring ferroptosis sensitivity. The healthy individual and patient information is summarized in Supplementary Table S3.
Plasma exosomes from healthy individuals (n = 20) and LUAD patients (n = 20) were used in 3D spheroids culture and the followed cell death analysis. The healthy individual and patient information is summarized in Supplementary Table S4.
Tissues from LUAD patients (n = 50) were used for analyzing cir93, correlations among intracellular cir93, NAT and FABP3. The patient information is summarized in Supplementary Table S5.
Tissues from LUAD patients (n = 30) were used for analyzing cir93 and FABP3 mutations. The patient information is summarized in Supplementary Table S6.
Plasma exosomes from healthy individuals (n = 100) and LUAD patients (n = 100) were used for analyzing NAT, acyl-CoA synthetase long-chain family member (ACSL4), lysophosphatidylcholine acyltransferase 3 (LPCAT3) and phospholipid transfer protein (PLTP). The healthy individual and patient information is summarized in Supplementary Table S7.
Plasma exosomes from healthy individuals (n = 15) and LUAD patients (n = 15) were used for analyzing correlations among exosomal, intracellular cir93 and FABP3. The patient information is summarized in Supplementary Table S8.
Tissues from LUAD patients (n = 82) were used for analyzing prognosis. The patient information is summarized in Supplementary Table S9.
2.4 Reagents and plasmids
The reagents erastin (Sigma), RSL3 (Sigma), GW4869 (Sigma), 5,5-(N-N-dimethyl)-amiloride hydrochloride (DMA, Sigma), deferoxamine (DFO, Sigma), Ferrostatin-1 (Fer-1, Sigma), NAT (Cayman, Ann Arbor, MI, USA), Actinomycin D (ActD, Sigma), C11-BODIPY581/591 (Invitrogen), propidium iodide (PI, Sigma), liproxstatin-1 (Lipro-1, Sigma), and salinomycin (Sigma) were used for cell treatments.
The wild-type (WT) and mutant cir93 (including cir93△R#1 and cir93△R#2) expressing plasmids were purchased from Geneseed Biotechnology (Guangzhou, Gongdong, China). Anti-cir93-1/2 plasmids were purchased from Sangon (Shanghai, China). WT-FABP3 and T cell leukemia homeobox 2 (TLX2) expressing plasmids were purchased from Biovision (Shanghai, China). Plasmids expressing mutant-FABP3, including FABP3△P#1, FABP3△P#2, FABP3△P#3, FABP3△P#4 and FABP3F16S, were constructed using overlapping PCR and cloned into pcDNA3.1(+) plasmids (Biolink, Shanghai, China). Constructs purchased from Geneseed Biotechnology were used for cir93 knockout, and lentiCRISPR v2-based constructs were used for FABP3 knockout. Short hairpin RNAs (shRNAs) targeting TLX2, transcription factor AP-2 alpha (AP-2α), IKAROS family zinc finger 1 (IKZF1), IKAROS family zinc finger 2 (IKZF2), heart and neural crest derivatives expressed 1 (HAND1), cysteine sulfinic acid decarboxylase (CSAD), and cysteine dioxygenase type 1 (CDO1) were purchased from Biovision. The WT and mutant (without intact TLX2 motif) promoters of ACSL4, LPCAT3, and PLTP were cloned into the pGL4.21 plasmids (Promega, Madison, WI, USA). The sequences for primers, sgRNAs and shRNAs are summarized in Supplementary Table S10.
2.5 Immunofluorescence (IF), IHC, immunoblotting (IB) and enzyme-linked immunosorbent assay (ELISA)
For IF, Paul Karl Horan-67 (PKH67) (1:500; #PKH67GL; Sigma) was probed to mark exosome packaged in A549/H1299 cells. Briefly, the exosome from A549 or H1299 cells expressing cir93-mCherry was mixed with Diluent C and PKH67 dye for 5 min and isolated using three sequential centrifugation steps as mentioned below. Then the isolated exosome was incubated with H1299 or A549 cells for 48h. Next, cells were harvested, fixed and blocked. Nuclei were counter- stained with DAPI. All images were collected via a confocal microscope (Leica, wetzlar, German).
For IHC, the primary antibodies used in the study included anti-alpha smooth muscle actin (αSMA; 1:100; #ab7817; Abcam, Hong Kong, China) and anti-4-HNE (1:100; #ab48506; Abcam). Briefly, the tissue sections were firstly deparaffinizated and rehydrated. Then, antigen retrieval was performed and endogenous peroxidases were blocked. Next, tissues were blocked and incubated with primary antibodies. The signals were detected with the Vectastain ABC kit (Vector Labs, Burlingame, CA, USA).
For IB, the primary antibodies used were, anti-FABP3 (1:1000; #ab133585; Abcam), anti-GAPDH (1:2000; #ab181602; Abcam), anti-CSAD (1:1000; #ab91016; Abcam), anti-CDO1 (1:1000; #232699; Abcam), anti-ACSL4 (1:2000; #ab155282; Abcam), anti-LPCAT3 (1:1000; #ab232958; Abcam), anti-PLTP (1:1000; #ab134066; Abcam), anti-Myc (1:2000; #2276; Cell Signaling Technology [CST], Boston, MA, USA), anti-cluster of differentiation 63 (CD63; 1:1000; #ab271286; Abcam), anti-tumor susceptibility 101 (TSG101; 1:1000; #125011; Abcam), anti-ALG-2 interacting protein X (ALIX; 1:1000; #ab275377; Abcam), anti-cluster of differentiation 9 (CD9; 1:1000; #ab92726; Abcam), anti-TLX2 (1:1000; PA5-34554; Invitrogen) and anti-calnexin (1:1000; #ab133615; Abcam). Briefly, cell protein lysates were separated in sodium dodecyl sulfate (SDS) polyacrylamide gels and transferred to nitrocellulose membranes. Blots were then incubated with specific antibodies, and signals were detected using the reader from Bio-rad (Hercules, CA, USA).
For ELISA, FABP3 levels were measured using kits from Yingxin Biotech Ltd. (Shanghai, China) as per the manufacturer's instructions. Briefly, samples and horseradish peroxidase (HRP) labeled detection antibodies were added into the micropores, incubated and washed. The substrate 3,3′,5,5′-tetramethylbenzidine (TMB) is then added and incubated. The absorbance (optical density value) at 450 nm was measured with a microplate reader (Biotek, Winooski, VT, USA).
2.6 Measurements of metabolites
AA was measured using a kit from CUSABIO (Houston, TX, USA, #CSB-E09040h), and MDA (#ab118970) and taurine (#ab241040) were measured using kits from Abcam in accordance with the manufacturer's instructions. NAT was analyzed by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS). The high-performance liquid chromatograph (HPLC) system was an Agilent 1100 (Agilent, Chandler, Arizona, USA). The mass spectrometry (MS) system was an AB 6500+ triple quadrupole tandem MS detection system (AB Sciex, Boston, MA, USA) equipped with an electrospray ionization (ESI) source and Analyst 1.7.1 workstation (AB Sciex).
2.7 Quantitative reverse transcription-PCR (qPCR)
Total RNA was extracted using Trizol reagent (Ambion, Carlsbad, CA, USA) and reversely transcribed into cDNA using the PrimeScriptTM RT reagent kit (Takara, Dalian, Liaoning, China). For real-time qPCR, the SYBR premix Ex Taq (Takara) kit was used for detecting the relative levels of nucleoporin 107 (NUP107), ACSL4, LPCAT3, PTLP, TLX2, AP-2α, IKZF1, IKZF2 and HAND1 mRNA, cir93 and circRNA_100934 (cir34). The reaction condition was 50°C for 3 min 95°C for 3 min, then 40 cycles of 95°C for 3 s and 60°C for 30 s. Semi-qPCR was also performed to examine NUP107 mRNA and cir93, and the reaction was terminated at the cycle 29, and the products were visualized by agarose gel electrophoresis. The primers are listed in Supplementary Table S10.
2.8 Measurements of cell viability, cell death, lipid reactive oxygen species (ROS) generation, ex vivo tissue slice culture and cell proliferation
Cell viability was detected using a CellTiter-Glo luminescent cell viability assay kit (Promega). Cell death was measured by staining with SYTOXTM green, followed by flow cytometry. Lipid ROS generation was analyzed by probing with C11-BODIPY581/591 at a final concentration of 1.5 μmol/L for 20 min before subjecting for further analysis using a flow cytometer. Ex vivo tissue slices were prepared as described in our previous study [27]. Afterward, DMSO, erastin or Fer-1 was co-incubated before tissue slices were stained with PI and photographed under a microscope. Because cell death can be measured directly and indirectly using the cell death and cell viability assays described above, we performed both to examine ferroptosis. Cell proliferation was measured using a Cell Counting Kit-8 (Beyotime).
2.9 Transmission electron microscope (TEM) analysis
For exosome observation, exosome was resuspended in 4% paraformaldehyde (PFA), and images were captured using the JEM1230 TEM system (JEOL, Tokyo, Japan). For morphological observation of mitochondria, cells were seeded onto 4-well chambered cover glass (Thermo Scientific, Waltham, MA, USA) at a density of 15,000 cells/well. Images were captured using the Olympus EM208S TEM (HITACHI, Tokyo, Japan).
2.10 Fluorescence in situ hybridization (FISH)
The main procedure was similar to that of conventional IHC but included several additional steps before adding primary antibodies. Briefly, tissues were incubated with a formamide/saline sodium citrate solution at 37°C for 4 h prior to the addition of digoxigenin (DIG)-labeled cir93 or cir34 probes (Sangon, listed in Supplementary Table S10) at 37°C overnight. Afterward, tissues were incubated with primary anti-DIG antibodies (1:200; #ab420; Abcam) and horseradish peroxidase-labeled secondary antibodies (CST, #8125) and signals were detected with the Vectastain ABC kit (Vector Labs).
2.11 Isolation and measurement of exosome
Exosome was isolated from cell cultured media or human plasma via three sequential centrifugation steps at 4°C: (1) 15 min at 500 × g to remove cells, (2) 30 min at 10,000 × g to remove cell debris and (3) ultracentrifugation at 110,000 × g for 70 min to pellet exosome. The pellet was finally re-suspended in phosphate-buffered saline (PBS) and centrifuged at 110,000 × g for another 70 min to remove soluble and secreted proteins. The concentration and size of exosome was analyzed using a NanoSight NS 300 system (NanoSight Technology, Malvern, UK).
2.12 Click chemistry for alkyne labeling
Cells were incubated with DMEM containing 10% FBS and 3% bovine serum albumin (BSA) with or without AA-alkyne or AdA-alkyne (20 mmol/L, Wuxi AppTec, Wuxi, Jiangsu, China) for 2 h. Next, the treatment medium was replaced by fresh DMEM containing 3% BSA for another 10 h, and cells were subsequently fixed in 4% PFA and permeabilized with 0.1% Triton-X in PBS. Afterward, cells were incubated with the PBS-based click reaction buffer (0.1 mmol/L Azide-fluor 488 [Cat# 760765, Sigma], 1 mmol/L CuSO4, 1 mmol/L tris(2-chloroethyl) phosphate, TCEP) for 1 h in a light-impermeable humidified chamber. Finally, cells were washed five times with 1 × PBS and blocked with PBS-BT (1 × PBS, 3% BSA, 0.1% Triton X-100, 0.02% NaN3) for 45 min at 25°C, followed by probing with concanavalin A-Alexa Fluor 350 and DAPI (Molecular Probes, Eugene, OR, USA). Images were captured using a confocal microscope (Leica).
2.13 Protein and RNA interaction analysis, RNA pull-down, RNA immunoprecipitation (RIP), and photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP)
The molecular basis underlying FABP3 and cir93 interactions was predicted by catRAPID (http://service.tartaglialab.com/page/catrapid_group) [28]. FABP3 and cir93 sequence were input into the corresponding box, the submit button was clicked and the prediction results would be displayed in a new window.
For RNA pull-down assays, synthesized cir93 (including WT and mutants) and its antisense probes labeled with biotin were ordered from Sangon (listed in Supplementary Table S10). Approximately 1 × 107 cells were lysed and incubated with 3 μg biotinylated probes at 4°C overnight. The biotin-coupled RNA-protein complex was pulled down with streptavidin magnetic beads (Life Technologies, Carlsbad, CA, USA) for another 4 h. After washing five times with PBS, the streptavidin beads were boiled, and the elutes were subjected to IB or proteomic analysis.
For RIP assays, a Magna RIP Kit (Merck Millipore, Billerica, MA, USA) was used. Briefly, cell lysates were incubated with magnetic beads loaded with 5 μg anti-FABP3 antibodies (1:100; #ab133585; Abcam) or control IgG (1:100; #3900; CST) overnight at 4°C. The remaining RNA after proteinase K digestion was extracted by TRIzol regent and analyzed by qPCR.
For PAR-CLIP, cells were incubated with (4-thiouridine, 4-SU, 250 μmol/L, Sigma) for 16 h followed by irradiation with 365 nm ultraviolet (UV) light for crosslinking. Subsequently, cells were lysed with NP40 lysis buffer on ice and centrifuged at 18,000 × g for 15 min to collect the supernatant, which was further incubated with 600 μL protein A Dynabeads (Invitrogen) bound with 15 μg anti-FABP3 antibodies (1:100; #ab133585; Abcam) for 2 h. After washing three times with IP wash buffer, beads were re-suspended and boiled at 95°C for 10 min. To detect FABP3-bound circRNAs, RNA was recovered and subjected to qPCR analysis.
2.14 Proteomic and metabolomic analyses
Proteomic and metabolomic analyses were performed, and the data were analyzed by Luming Biotechnology (Shanghai, China). Briefly, for the proteomic analysis regarding the proteins that were pulled down by cir93, the sample was separated by Nano-HPLC (EASY-nLC1200; Thermo Scientific) liquid phase system, and then analyzed by Q-Exactive MS (Thermo Scientific). For the direct proteomic analysis, the samples were labeled by tandem mass tag (TMT, Thermo Scientific), separated by Nano-HPLC (EASY-nLC1200; Thermo Scientific), and then analyzed by Q Exactive HF-X (Thermo Scientific). For the metabolomic analysis, the samples were separated and analyzed by a 7890B-5977A GC-MS (gas chromatography-mass spectrometry, Agilent, Wilmington, DE, USA). The proteomics data regarding the proteins that were controlled and pulled down by cir93 were deposited in the ProteomeXchange Consortium.
2.15 Promoter analysis, luciferase reporter assay, and chromatin immunoprecipitation (ChIP)
Transcription factors were predicted by LASAGNA-Search 2.0 (https://biogrid-lasagna.engr.uconn.edu) [29]. Briefly, we selected the Vertebrates transcription factor models, and input the promoter sequence in FASTA format, then clicked the Start Searching Button. The results could be summarized in a new window.
Firefly luciferase reporter plasmids for the detection of WT an mutant promoters of ACSL4, LPCAT3 and PLTP were co-transfected with Renilla luciferase reporter plasmids. After transfection, the ratios between firefly and Renilla luciferase activities were measured using a Dual-luciferase reporter assay system (Promega).
ChIP experiments were performed using the kits purchased from Active Motif (Carlsbad, CA, USA). Briefly, cells (2 × 107) were fixed using 1% formaldehyde, washed with PBS and lysed using lysis buffer. After sonication, protein-DNA complexes were incubated with antibody-coupled protein G beads at 4°C overnight. The next day, DNA was eluted in 1% SDS/0.1mol/L NaHCO3, reversed cross-link at 65°C, purified via phenol/chloroform extraction and ethanol precipitation and subjected to PCR analysis via agarose gel electrophoresis. The primary antibodies used were anti-TLX2 (1:50; PA5-34554; Invitrogen) and IgG (1:50; #3900; CST).
2.16 Statistical analysis
Tests used to examine the differences between groups included student's t-test, one-way and two-way analysis of variance (ANOVA) and χ2 test. The correlation between two groups was evaluated by the Spearman's rank-correlation analysis. The survival rate was calculated by the Kaplan-Meier method, and statistical significance was assessed by log-rank tests. A P < 0.05 was considered statistically significant.
3 RESULTS
3.1 Exosome antagonized lipid peroxidation and desensitized ferroptosis in LUAD
Excessive lipid peroxidation is a hallmark of ferroptosis, and we wondered if lipid peroxidation is suppressed in LUAD. MDA is an important byproduct of lipid peroxidation, and MDA levels were indeed lower in LUAD tissues compared with the matched adjacent normal tissues (Figure 1A). In addition, 39.9% of LUAD samples (75/188) showed the downregulation of 4-HNE, another byproduct of lipid peroxidation (Figure 1B). LUADs are frequently driven by mutations [3, 4]; however, after screening common mutations within EGFR (Del19, L858R, and T790M), kRAS (G12C, G12S, G12A, and G13C) and TP53 (R196P, H179Y, P250L, R249S, and R248G), we found that MDA was not likely regulated by these driver mutations (Figure 1C). Collectively, lipid peroxidation was suppressed in a large proportion of LUAD samples, possibly via a driver mutation-independent manner.
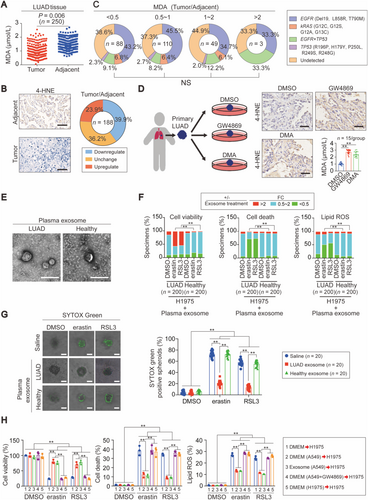
We next tested whether exosome suppresses lipid peroxidation and desensitizes LUAD cells to ferroptosis. Ex vivo primary LUADs from patients after curative therapy were treated with or without GW4869 and DMA, two exosome biogenesis inhibitors, and they both had elevated 4-HNE (Figure 1D), indicating that the prevention of exosome generation stimulates lipid peroxidation. Next, we asked whether exosome from LUAD plasma desensitize ferroptosis. To this end, plasma exosome from healthy individuals and LUAD patients was extracted and verified by TEM (Figure 1E). The exosome diameters were mainly between 30 and 150 nm (Supplementary Figure S1A), similar to those reported in prior studies [30, 31]. Except for the endoplasmic reticulum biomarker calnexin, exosome biomarkers including CD63, TSG101, ALIX and CD9 were all clearly detected in our plasma samples (Supplementary Figure S1B), further demonstrating the quality of the exosome. After testing several 3D spheroid cultured LUAD cell lines, A549 and H1975 cells were picked up for further study because they exhibited the lowest and the highest sensitivity to the two ferroptosis agents erastin and RSL3 (Supplementary Figure S1C-D). After pre-co-incubating mono-layer cultured H1975 cells with plasma exosome, we found that plasma exosome from a large proportion of LUAD patients could significantly prevent erastin- and RSL3-reduced cell viability and -induced cell death and lipid ROS generation (Figure 1F). In contrast, plasma exosome from healthy individuals had no significant effects (Figure 1F). The results from 3D spheroid cultures also supported that only the exosome from LUAD plasma had the capacity to prevent erastin- and RSL3-induced cell death in H1975 spheroids (Figure 1G). Because the sensitivity to ferroptosis varied between A549 and H1975 cells, we wondered whether the exosome from A549 cells mediated the reduced sensitivity of H1975 cells to ferroptosis. As expected, pre-co-incubation with exosome extracted from A549 cells cultured in DMEM or DMEM itself desensitized H1975 cells to the induction of erastin and RSL3 (Figure 1H). However, fresh DMEM or DMEM from cultured H1975 cells and A549 cells following GW4869 treatment did not have these effects (Figure 1H). Together, the role of exosome in antagonizing lipid peroxidation and desensitizing LUAD cells to ferroptosis was established.
3.2 The elevation in intracellular cir93 was closely linked with exosomal cir93 in LUAD
Next, the important components in exosome were investigated. Emerging studies have demonstrated the critical roles of exosomal circRNAs in tumorigenesis [32, 33]. Therefore, we aimed to identify important circRNAs in LUAD exosome. First, the available circRNA ChIP data (Zhu, et al. [34] and Chen, et al. [35] [accession No.: GSE101586]) on differential circRNA expression between LUAD and normal lung samples were analyzed. Two circRNAs, cir93 and cir34, were identified as the common upregulated intracellular circRNAs in LUAD in the two datasets (Figure 2A). Further analyses indicated that only cir93, but not cir34, was elevated in our LUAD cohort (n = 250, Figures 2B and 2C and Supplementary Figure S2A-B). Therefore, we focused on cir93 in subsequent experiments. We also confirmed cir93 as a true circRNA, because dislike NUP107 mRNA (the transcript of cir93 parental gene), cir93 could only be amplified by PCR from the cDNA but not the gDNA template of A549 and H1299 cells using circRNA-specific primer sets (Supplementary Figure S2C). Of note, elevated plasma exosomal cir93 was also observed in LUAD (n = 200) compared with healthy individuals (n = 200, Figure 2D). Because circRNAs are transferred between host and target cells via exosome [36], we suggested that the elevation in intracellular cir93 was closely linked with exosomal cir93 in LUAD.
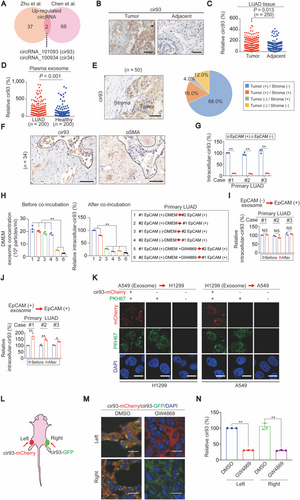
3.3 Exosome secreted by tumor cells was essential for increasing intracellular cir93 in LUAD
Because tumors and their stroma are both important for tumorigenesis [37, 38], we aimed to determine whether they are involved separately or cooperatively in mediating the link between exosomal and intracellular cir93. After measuring cir93 in LUAD specimens, we found that 68.0% (34/50) exclusively expressed cir93 in the tumor area, while only 4.0% (2/50) exhibited specific cir93 expression in the stroma (Figure 2E). The specimens with cir93 exclusively expressed in the tumor area were further evaluated by IHC using antibodies against αSMA, which is a biomarker of fibroblasts and to some extent capable of distinguishing the stroma from tumor areas. Indeed, all of the 34 samples showed mutual exclusion of cir93 and αSMA from the same areas (Figure 2F). EpCAM is a surface pan-carcinoma antigen. By sorting patient-derived EpCAM (+) and EpCAM (-) cells in primary tumors from three distinct LUAD patients, we found that intracellular cir93 was considerably higher in EpCAM (+) cells than in EpCAM (-) cells (Figure 2G). Taken together, the tumor area is more likely responsible for establishing the link between exosomal and intracellular cir93 in LUAD.
Subsequently, we aimed to investigate whether exosome secreted by tumor cells themselves elevates intracellular cir93 in the tumor area of LUAD. To this end, primary EpCAM (+) and EpCAM (-) cells from cases #1 and #2 in Figure 2G were used for further analysis. Before co-incubating cultured DMEM from the first cells with the secondary cells (The treatment methods of each group were listed in the box on the right side of Figure 2H), the exosome concentrations in cultured DMEM were confirmed to be similar between EpCAM (+) and EpCAM (-) cells (Figure 2H; lanes 1-4). However, intracellular cir93 in case #2 EpCAM (+) cells could only be maintained after co-incubating with the cultured DMEM from case #1 EpCAM (+) cells, and vice versa (Figure 2H; lanes 1-2). In contrast, in both cases #1 and #2, co-incubation with cultured DMEM from EpCAM (-) cells failed to maintain intracellular cir93 in EpCAM (+) cells (Figure 2H; lanes 3-4). The critical roles of exosome in EpCAM (+) cells were further verified because a reduction in exosome in the cultured DMEM of the first EpCAM (+) cells following GW4869 treatment failed to maintain intracellular cir93 in the secondary EpCAM (+) cells (Figure 2H; lanes 5-6). In addition to DMEM, the results from extracted exosome also demonstrated that only the exosome from EpCAM (+) but not EpCAM (-) cells had the capacity to elevate intracellular cir93 in EpCAM (+) cells (Figures 2I and 2J). Exosome-mediated cir93 transfer between A549 and H1299 cells was further confirmed by incubating one cell line with PKH67-labeled exosome extracted from another cell line expressing mCherry-labeled cir93, followed by tracing the exosome (green) and cir93 (red) under a microscope (Figure 2K). Together, exosome secreted by tumor cells themselves was essential for elevating intracellular cir93 in the tumor area of LUAD.
To further confirm that LUAD cells directly produce cir93 and simultaneously obtain cir93 from exosome in vivo, CDXs were generated from A549 cells exclusively expressing either mCherry (red)- or green fluorescent protein (GFP, green)-labeled cir93 from the left and right sides of mice, respectively (Figure 2L). Overleaping signals (yellow) were detected in CDXs at both sites, which were blocked by administrating GW4869 (Figure 2M), suggesting that LUAD cells obtain cir93 from each other via exosome. Moreover, the concentrations of cir93 were significantly reduced (Figure 2N). Because there was no other exogenous source of cir93, our data strongly suggest that LUAD cells maintained cir93 expression by generating cir93 and providing cir93 via exosome.
3.4 Elevated intracellular cir93 desensitized LUAD cells to ferroptosis via regulating AA
Although enhanced intracellular cir93 levels are maintained by exosome in LUAD (Figure 2), whether exosome antagonizes ferroptosis via elevating intracellular cir93 remains unclear. Ferroptosis is characterized by the presence of smaller than normal mitochondria with condensed mitochondrial membrane densities [39]. As expected, treating H1975 cells with erastin and RSL3 resulted in these effects; however, they were prevented by overexpressing cir93 (Figure 3A). By pre-infecting H1975 cells with a dual cir93- and GFP-expressing lentivirus and staining the cells with PI (a small fluorescent molecule to detect dead cells), elevating cir93 was also revealed to prevent erastin- and RSL3-induced cell death (Figure 3B). In addition, similar to the ferroptosis inhibitors Fer-1 and DFO, overexpressing cir93 prevented the erastin- and RSL3-induced reduction in cell viability and increase in lipid ROS generation in H1975 cells (Figure 3C and Supplementary Figure S3A). Interestingly, cir93 itself was not affected by erastin and RSL3 (Supplementary Figure S3B), suggesting that cir93 was an independent factor that influences ferroptosis. If cir93 desensitizes LUAD cells to ferroptosis, then anti-cir93 should exert the opposite function. As expected, anti-cir93 administration downregulated intracellular cir93 in A549 cells (Supplementary Figure S3C), and aggravated the erastin- and RSL3-induced reduction in cell viability and increase in cell death and lipid ROS generation (Supplementary Figure S3D-F). Thus, the above data demonstrated the role of elevated cir93 in mitigating ferroptosis in vitro.
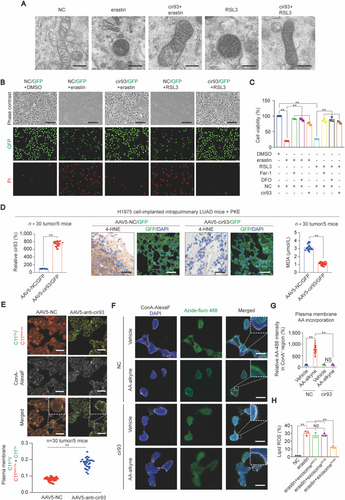
To investigate whether elevating intracellular cir93 also desensitizes LUAD cells to ferroptosis in vivo, H1975 cell-implanted intrapulmonary LUAD-bearing mice were intranasally infected with an AAV5 dual-expressing cir93 and GFP before being administered with PKE, an in vivo stable erastin derivative. Compared with the control, cir93 was indeed overexpressed, and PKE-induced elevations in 4-HNE and MDA were largely prevented in intrapulmonary LUAD mice (Figure 3D), suggesting that enhancing cir93 expression suppressed ferroptosis-associated lipid peroxidation in vivo. To further investigate whether anti-cir93 stimulates lipid peroxidation within plasma membrane fractions in mouse LUAD, the fluorescent probe C11-BODIPY581/591 was used to directly measure oxidized lipids, and concanavalin A-Alexa Fluor 350 was applied to visualize membranes. Images were captured with an emission filter at 580/600 nm (the non-oxidized form, red) and 490/510 nm (the oxidized form, green) and then merged to visualize the fraction of oxidized C11-BODIPY581/591. As shown in Figure 3E, the fraction of oxidized C11-BODIPY581/591 (green) within the plasma membrane was remarkably increased following the administration of anti-cir93. Combined with the data showing that overexpressing cir93 desensitizes cells to ferroptosis induced by both erastin and RSL3 (Figure 3A-C), and because treatment with two small molecules both led to lipid peroxidation [40], we supposed that increasing intracellular cir93 might desensitize LUAD cells to ferroptosis via suppressing lipid peroxidation.
Next, we investigated how does elevating intracellular cir93 suppress lipid peroxidation in LUAD cells. Excessive peroxidation of PUFAs, such as AA and AdA, is a prerequisite for the activation of ferroptosis [26]. Plasma membrane incorporation of AA is also essential for this process [41]. Using a click chemistry-based method with alkyne-labeled AA or AdA and Fluro-488-labeled Azide, we found that elevating cir93 in H1975 cells reduced AA but not AdA incorporation into the plasma membrane (Figures 3F and 3G and Supplementary Figure S3G-H). These results suggested that suppression of lipid peroxidation by elevated cir93 might be mediated through its modulation of AA.
Finally, we examined whether exosome suppresses lipid peroxidation in a cir93-dependent manner. Because MRC-5 and WI-38, two established lung fibroblast cell lines, demonstrated remarkably lower levels of intracellular cir93 and exosomal cir93 in cultured DMEM compared with LUAD cell lines (Supplementary Figure S3I), we compared the effects of exosome extracted from MRC-5, WI-38 and LUAD A549 cells on lipid ROS generation following erastin treatment in H1975 cells. We observed that only exosome from A549 cells significantly reduced erastin-induced lipid ROS generation (Figure 3H), indicating that exosome suppressed lipid peroxidation in LUAD cells via a cir93-dependent mechanism.
3.5 cir93 interacted with and upregulated the downstream factor FABP3
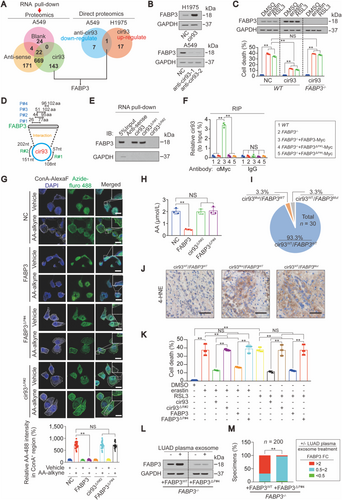
Then, the downstream effectors of cir93 were further investigated. Given that circRNAs interact with proteins [42, 43], we performed proteomics to identify potential proteins influenced by cir93. Using pull-down experiments with cir93, anti-sense cir93 or a blank control (without RNA), followed by proteomics, 143 proteins specifically interacting with cir93 were identified (Figure 4A). To narrow down these candidates, we determined which proteins are also regulated by cir93. Because intracellular cir93 was higher in A549 cells than in H1975 cells (Supplementary Figure S3I), we inhibited cir93 in A549 cells and overexpressed cir93 in H1975 cells before re-subjecting samples to proteomics analysis. Only FABP3 was identified to be upregulated by cir93. Unfortunately, no proteins were identified to be downregulated by cir93. Interestingly, FABP3 was also one of the 143 proteins found to interact with cir93 (Figure 4A). The upregulation of FABP3 by cir93 was further verified by IB (Figure 4B). Similar to cir93, FABP3 was not regulated by erastin and RSL3 (Figure 4C), indicating that FABP3 is also an independent factor that influences ferroptosis. Moreover, FABP3 was indispensable for the elevated cir93-induced desensitization of LUAD cells to erastin- and RSL3-induced ferroptosis and ferroptosis-associated lipid ROS generation because compared with WT H1975 cells, cir93 was ineffective in FABP3−/− cells (Figure 4C and Supplementary Figure S4A-C). Together, our findings suggested that FABP3 was a downstream target of cir93. Moreover, cir93 desensitized LUAD cells to ferroptosis by upregulating the expression of FABP3.
3.6 Molecular basis behind cir93-FABP3 interaction and its essential role in modulating FABP3, AA, lipid peroxidation, and sensitivity of LUAD cells to ferroptosis
Subsequently, we investigated the specific regions of cir93 required for its ability to interact with and upregulate FABP3. To further verify that FABP3 interacts with cir93, PAR-CLIP was performed, and we found that FABP3 interacted with cir93 but not cir34 (Supplementary Figure S4D). The molecular basis underlying FABP3 and cir93 interactions was then predicted by the online software catRAPID. Three regions, named as P#1 (26th∼77th a.a), P#2 (44th∼95th a.a) and P#3 (51st∼102nd a.a), located within FABP3 and two regions, named as R#1 (57th∼108th nt) and R#2 (151st ∼202nd nt), located within cir93 were predicted with interaction potentials (Figure 4D and Supplementary Figure S4E). cir93 pull-down experiments demonstrated that deletion of the R#2 region of cir93 completely abolished cir93-FABP3 interactions in H1975 cells (Figure 4E). Reconstitution of FABP3−/− H1975 cells by Myc-tagged WT or FABP3 without P#3 region followed by RIP experiments using anti-Myc antibodies indicated that the P#3 region of FABP3 contains a critical domain responsible for the cir93-FABP3 interaction (Figure 4F). The P#1 and P#2 regions of FABP3 were excluded because FABP3 still interacted with cir93 even when they were deleted (Supplementary Figure S4F). Because P#1, P#2, and P#3 had overlapping regions and only the region containing the 96th∼102nd aa in P#3 was unique as compared with P#1 and P#2 (Figure 4D). Therefore, we speculated that this region (hereafter named as P#4) participates in the cir93-FABP3 interactions. As expected, deletion of P#4 disrupted cir93-FABP3 interactions in H1975 cells (Figure 4F). Furthermore, in addition to P#3, P#4 was required for cir93 to upregulate FABP3 (Supplementary Figure S4G). Taken together, the molecular basis for cir93-FABP3 interactions and the specific regions required for upregulating FABP3 in LUAD cells were elucidated.
We next asked whether cir93-FABP3 interactions also influence AA, lipid peroxidation and LUAD cell sensitivity to ferroptosis. Similar to increasing cir93, overexpressing FABP3 reduced AA but not AdA incorporation into the plasma membrane of H1975 cells. However, the effects were no longer observed when the P#4 site was deleted. Deletion of the R#2 region also prevented the ability of cir93 to suppress AA membrane incorporation (Figure 4G and Supplementary Figure S4H-I). The data from Figure 4H further demonstrated that the cir93-FABP3 interactions were required for the reduction in global AA. To obtain supporting clinical data, intracellular cir93 and the open reading frame (ORF) of FABP3 were sequenced in 30 randomly selected LUAD tissue specimens. Only one specimen (3.3%) each contained a mutation in R#2 of cir93 and P#4 of FABP3, respectively (Figure 4I). We next verified that these two mutations disrupted cir93-FABP3 interactions (Supplementary Figure S4J-K). Of note, mutations in cir93 and FABP3 increased 4-HNE concentrations in LUAD (Figure 4J), further confirmed the importance of cir93-FABP3 interaction in reducing lipid peroxidation. Moreover, disruption of cir93-FABP3 interaction abolished the ability of cir93 and FABP3 to desensitize H1975 cells to erastin- and RSL3-induced cell death (Figure 4K). Collectively, cir93-FABP3 interactions and their upregulation of FABP3 were essential for modulating AA and reducing lipid peroxidation and LUAD cell sensitivity to ferroptosis.
3.7 Exosome upregulated FABP3 via cir93-FABP3 interactions in LUAD
Because the cir93-FABP3 interactions are required for cir93 to upregulate FABP3 (Figure 4B and Supplementary Figure S4G), and exosome is essential for elevating intracellular cir93 in LUAD (Figure 2), we wondered whether plasma exosome from LUAD cells upregulates intracellular FABP3 expression via the cir93-FABP3 interactions. To address this, plasma exosome from LUAD (n = 200) cells was co-incubated with engineered H1975 cells expressing WT FABP3 or FABP3 without P#4 region. 68.5% of (137/200) exosome upregulated FABP3 by more than two-fold in FABP3WT-expressing H1975 cells, whereas only 3.5% of (7/200) exosome exhibited similar functions in cells expressing FABP3△P#4 (Figures 4L and 4M), suggesting that cir93-FABP3 interactions were also required for exosome to upregulate FABP3 in LUAD.
3.8 Iron was not involved in FABP3-reduced AA incorporation into the plasma membrane
Because iron is essential for ferroptosis, we wondered if there is a relationship between changes in iron and FABP3-mediated AA transportation. To address this, salinomycin, a ferroptosis inducer that activates ferritin degradation and increases the labile iron level [44], was used in H1975 cells with or without FABP3 overexpression. The ability of salinomycin to elevate labile iron was verified in H1975 cells (Supplementary Figure S4L). However, FABP3-reduced AA incorporation into the plasma membrane was unaffected by salinomycin (Supplementary Figure S4M-N). Thus, we concluded that iron is not involved in FABP3-reduced AA transportation.
3.9 Taurine was essential for cir93-mediated upregulation of FABP3 to modulate AA and desensitize LUAD cells to ferroptosis
Next, we investigated the effects of FABP3 upregulation in LUAD. Because FABP3 is required for AA transport and metabolism [45, 46], metabolites regulated by FABP3 and associated with AA might determine the sensitivity of LUAD cells to ferroptosis. To identify these metabolites, metabolomics analyses were performed to compare metabolites in LUAD tissues expressing high and low levels of FABP3. Among 362 metabolites, 63 were upregulated, and 26 were downregulated in high FABP3-expressing LUADs (n = 30) compared with the low-expressing samples (n = 30, Figure 5A and Supplementary Figure S5A). To the best of our knowledge, among all the identified metabolites, taurine was the only one that can also react with AA to generate NAT [47]. Because taurine and AA are substrates, and NAT is a product of this reaction (Figure 5B), the reduced taurine in high FABP3-expressing LUAD samples (Figure 5A) might be explained by the FABP3-mediated acceleration of the reaction. Unfortunately, no decrease in global AA or increase in NAT was detected by metabolomics, which might be because of technical limitations [48]. However, by ELISA and targeted MS analyses, we found that global AA was reduced and NAT was induced following FABP3 overexpression in H1975 cells (Supplementary Figure S5B-C). FABP3 transports AA via its phenylalanine (F) 16 residue [46]. Hence, we replaced this F residue with serine (S) to test whether the reduced taurine is mediated by the transport function of FABP3. As expected, overexpressing FABP3F16S failed to reduce taurine as compared with FABP3WT in H1975 cells (Figure 5C). These results suggested that FABP3 upregulation stimulated AA transport and its subsequent reaction with taurine, thereby reducing global AA in LUAD cells.
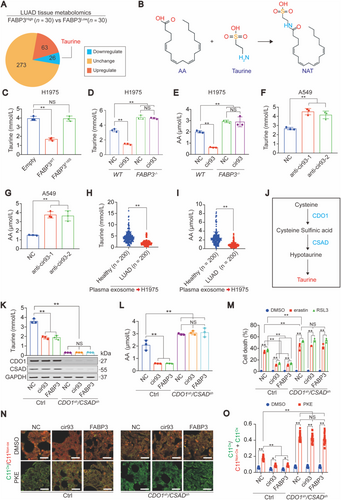
Because FABP3 works as a downstream factor of cir93 (Figure 4C and Supplementary Figure S4B-C), we wondered whether cir93 is also associated with FABP3-mediated changes in taurine and global AA. Overexpressing cir93 in H1975 cells led to simultaneously reduced taurine and global AA, and their levels were completely abolished by knocking out FABP3 (Figures 5D and 5E), demonstrating that cir93 regulates taurine and global AA via FABP3. In contrast, inhibiting cir93 via anti-cir93 in A549 cells simultaneously induced taurine and global AA at the same time (Figures 5F and 5G). Thus, manipulating taurine and global AA alterations are downstream effects of cir93 in LUAD cells. In addition, co-incubation of H1975 cells with plasma exosome from LUAD patients also resulted in lower levels of taurine and global AA as compared with the exosome from healthy individuals (Figures 5H and 5I). Given the importance of AA in ferroptosis, the exosome-induced desensitization of LUAD cells to ferroptosis might partially be mediated via a reduction in global AA in a cir93-FABP3-taurine-dependent manner.
To further verify the essential roles of taurine in reducing global AA and desensitizing LUAD cells to ferroptosis, we aimed to eliminate endogenous taurine in LUAD cells. CDO1 and CSAD are critical for the synthesis of taurine (Figure 5J). By simultaneously knocking down CDO1 and CSAD in H1975 cells, taurine was significantly reduced but could not be further decreased anymore by overexpressing cir93 or FABP3 (Figure 5K), suggesting that sufficient taurine was a prerequisite for cir93 and FABP3 to regulate taurine itself. Of note, taurine was indeed essential for cir93 and FABP3 to reduce global AA (Figure 5L). We then investigated whether taurine is indispensable for cir93 and FABP3 to desensitize LUAD cells to ferroptosis and reduce lipid ROS generation. Following the depletion of taurine, erastin- and RSL3-induced ferroptosis and lipid ROS generation were significantly aggravated at basal levels; however, the restoration effects mediated by cir93 and FABP3 were all inhibited (Figure 5M and Supplementary Figure S5D), suggesting that taurine was an anti-ferroptosis factor and essential for cir93 and FABP3 to desensitize LUAD cells to ferroptosis. By evaluating oxidized C11-BODIPY581/591 fractions and MDA concentrations in LUAD cells implanted in mice, the essential role of taurine in the cir93- and FABP3-mediation reduction in lipid peroxidation following PKE administration in vivo was further verified (Figures 5N and 5O and Supplementary Figure S5E).
3.10 NAT prevented AA incorporation into the plasma membrane in LUAD cells
As described above, we used a click chemistry-based method to evaluate AA incorporation into the plasma membrane. However, click chemistry was performed in the presence of excessive exogenous alkyne-labeled AA. Under these conditions, overexpressing cir93 and FABP3 still prevented AA incorporation into the plasma membrane, although endogenous global AA was also reduced by cir93 and FABP3 (Figures 5E, G, and L, Figures 6A and 6B and Supplementary Figure S6A), indicating that an additional mechanism might be simultaneously involved. Because AA consumption is accompanied by NAT generation (Figure 5B), we speculated that the nascent NAT might be essential for the function of cir93 and FABP3. As expected, the relationships among cir93, FABP3 and NAT were established in H1975 cells (Figures 6C and 6D). Additionally, taurine was essential for cir93 and FABP3 to elevate NAT in H1975 cells and implanted tumors (Figure 6E and Supplementary Figure S6B). Interestingly, compared with plasma exosome from healthy individuals, co-incubation with exosome from LUAD patients resulted in a higher level of intracellular NAT in H1975 cells (Figure 6F), further indicating that elevating NAT is a critical event for exosome to desensitize LUAD cells to ferroptosis. To provide direct evidence supporting the role of NAT in preventing AA incorporation into the plasma membrane, NAT was directly incubated with H1975 cells, and its function of NAT was confirmed (Figures 6G and 6H). Then, we investigated whether NAT desensitizes LUAD cells to ferroptosis, and found that erastin- and RSL3-induced induction of cell death and lipid peroxidation and reduction of cell viability were alleviated in the presence of NAT (Figures 6I and 6J and Supplementary Figure S6C-E). Thus, cir93- and FABP3-induced desensitization of LUAD cells to ferroptosis might also be mediated via the stimulation of NAT generation to prevent AA incorporation into the plasma membrane.
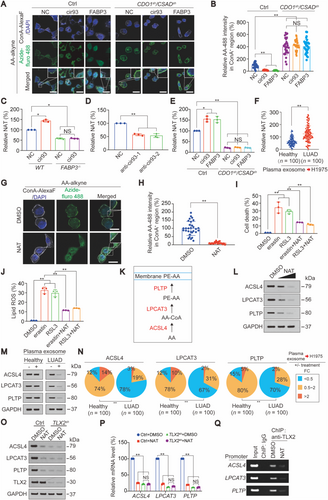
Next, we investigated the related mechanisms. As known, ACSL4, LPCAT3 and PLTP are enzymes involved in the incorporation of AA into the plasma membrane (Figure 6K) [49, 50]. Therefore, we tested whether NAT affected these enzymes, and observed that they were all dose-dependently reduced by NAT in H1975 cells (Figure 6L). By comparing fold changes in ACSL4, LPCAT3, and PTLP in the presence or absence of co-incubation with plasma exosome, we found that 78% (78/100), 67% (67/100) and 70% (70/100) of exosome from LUAD patients reduced ACSL4, LPCAT3, and PTLP, respectively, to a level less than 50%. However, similar effects were found for only 12% (12/100), 12% (12/100) and 15% (15/100) of exosome from healthy individuals (Figures 6M and 6N). Together, our data indicate that NAT-mediated downregulation of enzymes associated with AA incorporation into the plasma membrane is equally important for exosome, cir93 and FABP3 to desensitize LUAD cells to ferroptosis.
To further investigate how NAT suppresses ACSL4, LPCAT3, and PLTP expression, we performed bioinformatics to analyze the promoters of ACSL4, LPCAT3 and PLTP genes and revealed that TLX2, AP-2α, IKZF1, IKZF2, and HAND1 overlapped as common potential transcription factors that bind to these three promoters (Supplementary Figure S6F). We further observed that only when TLX2 was knocked down, the mRNA expression of all three genes was simultaneously reduced (Supplementary Figure S6G-H), suggesting that TLX2 generally stimulates ACSL4, LPCAT3 and PLTP gene expression. Additionally, in the absence of the TLX2-binding domain (Supplementary Figure S6I), the promoter activities of all three genes were significantly reduced, and the ability of NAT to suppress promoter activity was diminished (Supplementary Figure S6J). Moreover, we verified that NAT suppressed the protein and mRNA expression of ACSL4, LPCAT3, and PLTP via a TLX2-dependent mechanism (Figures 6O and 6P) and that NAT was capable of displacing TLX2 from promoters (Figure 6Q), indicating that NAT reduces ACSL4, LPCAT3, and PLTP expression via suppressing TLX2 binding to their promoters.
Because TLX2 is a key transcription factor that upregulates ACSL4, LPCAT3, and PLTP, we investigated whether TLX2 is required for cir93, FABP3, and NAT to desensitize H1975 cells to erastin- and RSL3-induced ferroptosis and lipid ROS generation. We found that TLX2 overexpression protected LUAD cells against these effects, whereas knocking out TLX2 further exacerbated these effects (Supplementary Figure S6K-M), demonstrating that TLX2 functions downstream of the cir93/FABP3/NAT axis to modulate the sensitivity of LUAD cells to ferroptosis.
3.11 Further verification of the role of cir93 in exosome to modulate ferroptosis
To further validate the role of cir93 in exosome, we collected exosome from cir93−/− A549 cells and confirmed that exosomal cir93 was almost undetectable in these exosomes (Supplementary Figure S6N). By incubating H1975 cells with exosome from A549 cells, we found that the ability of exosome to modulate lipid ROS generation, FABP3, ACSL4, LPCAT3 and PTLP expression, taurine, AA and NAT concentrations and AA incorporation into the plasma membrane were all diminished when cir93 was depleted (Supplementary Figure S6O-U). Thus, we further verified that the role of exosome in modulating ferroptosis in LUAD cells is cir93-dependent.
3.12 The correlations among exosomal and intracellular cir93, FABP3, taurine, and NAT in LUAD
To further verify the above conclusions from above cell- and mouse-based experiments (Figures 1-6), we evaluated correlations among exosomal and intracellular cir93, FABP3, taurine, and NAT in human LUAD specimens. Similar to intracellular cir93 (Figures 2B and 2C), FABP3 was also elevated in our LUAD cohort (n = 250, Supplementary Figure S7A). A significant correlation between FABP3 and intracellular cir93 was revealed in LUAD (n = 250, Figure 7A). Moreover, taurine was negatively correlated with intracellular cir93 and FABP3 (Figures 7B and 7C). In contrast, positive correlations among NAT, intracellular cir93 and FABP3 were identified in LUAD (Figures 7D and 7E). To further evaluate the close relationship between exosomal and intracellular cir93, matched plasma and tissue specimens were obtained from the same LUAD patients (n = 15), and we found that exosomal cir93 was positively correlated with intracellular cir93 and FABP3 (Supplementary Figure S7B-C). Therefore, the close relationships among exosomal and intracellular cir93, FABP3, taurine and NAT were established in LUAD.
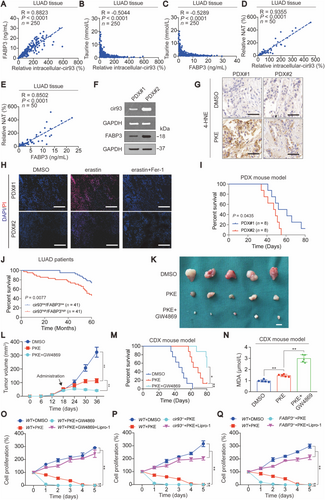
3.13 Ferroptosis resistance and poor survival outcome predicted by high levels of cir93 and FABP3 in LUAD
PDX mouse models are promising tools to evaluate drug efficacy in vivo. We compared the efficacy of PKE in two PDX mouse models expressing distinct levels of cir93 and FABP3 (Figure 7F), and observed that higher levels of cir93 and FABP3 in PDX#2 resulted in reduced suppression of tumor growth and lower 4-HNE levels following PKE administration compared with PDX#1 with lower levels of cir93 and FABP3 (Figures 7F and 7G and Supplementary Figure S7D). These findings further demonstrated that cir93 and FABP3 stimulate tumor growth and desensitize cells to ferroptosis in LUAD. By evaluating ex vivo tissue slices of PDX models, we also found that PDX#2 was more resistant to erastin-induced ferroptosis compared with PDX#1 (Figure 7H). Overall survival (OS) was subsequently evaluated for PDX mouse models, and PDX#2 demonstrated a shorter OS than PDX#1 (Figure 7I). Shorter OS was also observed in LUAD patients with higher levels of cir93 and FABP3 (n = 41) compared with those with lower levels (n = 41, Figure 7J). These data demonstrated that ferroptosis resistance and poor survival in LUAD were predicted by high levels of cir93 and FABP3.
3.14 Blocking exosome improved ferroptosis-based treatment
Because exosome from LUAD desensitizes cells to ferroptosis (Figures 1-6), we wondered whether blocking the biosynthesis of exosome improves ferroptosis-based treatment. To this end, we further co-administrated GW4869 to PKE-treated A549-based CDX mice. We found that co-treating mice with GW4869 more significantly impaired tumor growth and prolonged OS compared with PKE treatment alone (Figure 7K-M). The aggravated elevation in MDA further confirmed that these effects were mediated through a lipid peroxidation-dependent manner (Figure 7N). Together, our data suggest that co-treatment with agents that block the biosynthesis and function of exosome might help improve ferroptosis-based therapies in LUAD.
3.15 Ferroptosis was involved in the inhibition of exosome/cir93/FABP3-mediated tumor suppression
Finally, to demonstrate that ferroptosis plays a role in tumor suppression mediated by the inhibition of exosome/cir93/FABP3, we evaluated cell proliferation in A549 cells with or without co-treatment with Lipro-1, a ferroptosis inhibitor. We found that treating A549 cells with PKE impaired cell proliferation (Figure 7O-Q), which could be strengthened by co-treating with GW4869, or knocking out cir93 or FABP3. However, all these effects were blocked by Lipro-1 (Figure 7O-Q), confirming that ferroptosis is involved in tumor suppression following exosome/cir93/FABP3 inhibition in LUAD cells.
4 DISCUSSION
Not only intracellular system, extracellular system also has the ability to activate antioxidant function and inhibit ferroptosis. Prior studies suggested that exosome secreted from cancer-associated fibroblasts (CAFs) and mesenchymal stem cells (MSCs) in tumor microenvironment plays key roles in suppressing ferroptosis [21, 51]. In the present study, we revealed that exosome released from tumor cells increased intracellular cir93 to upregulate FABP3 and desensitize LUAD cells to ferroptosis via a FABP3-dependent reduction in global AA and prevention of AA incorporation into the plasma membrane. (Figure 8).
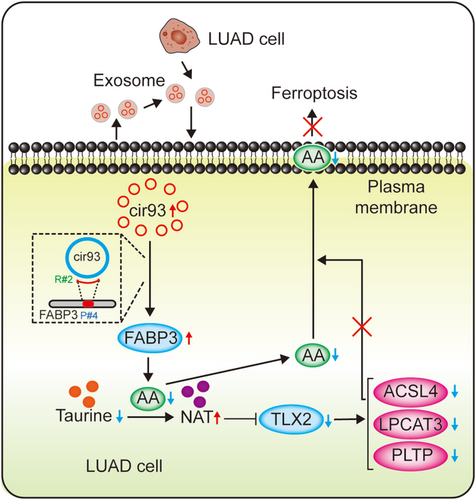
In addition to the most well-accepted function of ceRNAs as miRNA sponges [23], circRNAs also directly interact with proteins to perform physiological functions [42, 43]. In the present study, we added another example of this. Specifically, we showed that cir93-FABP3 interactions are required for the upregulation of FABP3 in LUAD cells. However, the mechanism by which cir93 elevates FABP3 remains unknown. Recently, circRNA cerebellar degeneration-related protein 1 transcript antisense was revealed to directly interact with p53 via its DNA-binding domain. Interestingly, this interaction also disrupts complex formation between p53 and its ubiquitin E3 ligase (E3) mouse double minute 2 homolog, thereby protecting p53 from degradation and increasing its expression [52]. Because cir93-FABP3 interactions are also critical for cir93 to increase FABP3 expression, cir93 might interfere with the ubiquitination of FABP3. However, the E3 involved remains unclear and needs to be identified and verified.
Phosphoethanolamine-containing AA is a key phospholipid that undergoes peroxidation and is essential for ferroptosis [26, 41]. Evidence demonstrates that supplementing AA sensitizes cells to ferroptosis [53]. However, the regulation of AA per se is not completely known. In this study, we elucidated that global AA was reduced through its reaction with taurine, and this process was further promoted by FABP3. FABP3 is a critical factor for PUFA transport, including AA [45, 46]. Our data supported that the reaction between AA and taurine is also indispensable for the transport function of FABP3. Given that FABP3 transports AA to proteins, such as α-synuclein [45], we hypothesize that some certain proteins might work as adaptors, providing reaction sites for AA and taurine. However, these proteins remain to be identified, which is an aim of our future study. Interestingly, the product of AA and taurine (i.e., NAT) prevents AA incorporation into the plasma membrane, thus further reducing the opportunity for PUFA peroxidation in the membrane. NAT reduces ACSL4, LPCAT3 and PLTP, which are all essential for AA incorporation into the plasma membrane [49, 50]. The transcription factor TLX2 is targeted by NAT to reduce ACSL4, LPCAT3 and PLTP promoter activities. Therefore, we also elucidated the molecular mechanism by which NAT reduces AA incorporation into the plasma membrane.
Cellular protection against oxidative damage in ferroptosis is organized by antioxidant systems. Antioxidants, such as glutathione, coenzyme Q10 and tetrahydrobiopterin, are critical for protecting tumor cells against ferroptosis [54]. Prior findings from our lab also confirmed that ferroptosis is suppressed by the maintained synthesis and utilization of glutathione [6, 7, 55, 56]. However, antioxidants that protect against ferroptosis are not completely known. Based on its role in reducing global AA and generating NAT, taurine is essential to prevent excessive lipid peroxidation, and thus can be regarded as a newly discovered antioxidant protecting against ferroptosis.
Recently, ferroptosis-based therapy has been recognized as a potential approach to treat cancer [8, 40]. The cancer-killing effect of sorafenib, a Food and Drug Administration (FDA, USA)-approved small molecule compound for cancer therapy, has been linked to ferroptosis [57]; other ferroptosis-related gene therapies and nanomaterials are currently being developed [58]. However, there are several limitations of ferroptosis-based therapy that need to be addressed before it is applied in clinical settings, including its off-target effects, unsatisfactory efficacy, unguaranteed safety and low in vivo stability of inducers [8, 40, 59]. Here, we provide several new ferroptosis targets including cir93, FABP3, taurine and NAT, and their functions are all associated with exosome. Indeed, the efficacy of ferroptosis was enhanced by blocking exosome biosynthesis in our in vivo preclinical models, thereby providing a new strategy to improve ferroptosis-based therapy.
5 CONCLUSIONS
Exosome and cir93 are critical to desensitize LUAD to ferroptosis. Blocking exosome might help enhance the efficacy of ferroptosis-based therapy in future treatments for LUAD.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Ethics Committee of Shanghai Chest Hospital (permit number: K19-11). Each participant signed an informed consent before participating to this study. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of Shanghai Chest Hospital (permit number: KS [Y] 21028).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets generated and/or analyzed during the current study are available in the ProteomeXchange Consortium under the accession number PXD025491 (username: [email protected]; password: U8S9cjhj) and PXD025575 (username: [email protected]; password: 9h0oFtLK).
ACKNOWLEDGEMENTS
We thank Melissa Crawford from Liwen Bianji (Edanz; www.liwenbianji.cn/), for editing the English text of this manuscript. This work was supported by the National Natural Science Foundation of China (81871907, 81822029, 81872288, 82173015, 81902315, 81902869, and 81774291), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine (20191834), Project of Clinical Research Supporting System, Clinical Medicine First-class Discipline, Grant Support “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (18CG16), Shanghai Sailing Program (19YF1444800), Science and technology commission of Shanghai municipality project (19140902600 and 21140902800), and Nurture projects for basic research of Shanghai Chest Hospital (2018YNJCM01, 2019YNJCM06, 2021YNZYJ01, 2021YNZYY01, and 2021YNZYY02).
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS' CONTRIBUTIONS
X.Z. analyzed the data and wrote the manuscript. Y.X. and K.Y. collected and analyzed the clinical samples and data. L.M., X.X. (Xin Xu), and F.M. analyzed the data. Y.N. designed and organized the images. Y.S. performed the bioinformatics analysis. S.G., X.X. (Xiangfei Xue), Y.W., S.Q., J.C. and H.W. analyzed and discussed the data. X.T. constructed the plasmids. Y.M. performed the mouse experiments. Y.Q. and Y.Y. designed the study. J.W. designed the study and wrote the manuscript. All authors read and approved the final manuscript.




