Increased mitochondrial fission drives the reprogramming of fatty acid metabolism in hepatocellular carcinoma cells through suppression of Sirtuin 1
Dan Wu and Yi Yang contributed equally to this work.
Abstract
Background
Mitochondria are dynamic organelles that constantly change their morphology through fission and fusion processes. Recently, abnormally increased mitochondrial fission has been observed in several types of cancer. However, the functional roles of increased mitochondrial fission in lipid metabolism reprogramming in cancer cells remain unclear. This study aimed to explore the role of increased mitochondrial fission in lipid metabolism in hepatocellular carcinoma (HCC) cells.
Methods
Lipid metabolism was determined by evaluating the changes in the expressions of core lipid metabolic enzymes and intracellular lipid content. The rate of fatty acid oxidation was evaluated by [3H]-labelled oleic acid. The mitochondrial morphology in HCC cells was evaluated by fluorescent staining. The expression of protein was determined by real-time PCR, iimmunohistochemistry and Western blotting.
Results
Activation of mitochondrial fission significantly promoted de novo fatty acid synthesis in HCC cells through upregulating the expression of lipogenic genes fatty acid synthase (FASN), acetyl-CoA carboxylase1 (ACC1), and elongation of very long chain fatty acid protein 6 (ELOVL6), while suppressed fatty acid oxidation by downregulating carnitine palmitoyl transferase 1A (CPT1A) and acyl-CoA oxidase 1 (ACOX1). Consistently, suppressed mitochondrial fission exhibited the opposite effects. Moreover, in vitro and in vivo studies revealed that mitochondrial fission-induced lipid metabolism reprogramming significantly promoted the proliferation and metastasis of HCC cells. Mechanistically, mitochondrial fission increased the acetylation level of sterol regulatory element-binding protein 1 (SREBP1) and peroxisome proliferator-activated receptor coactivator 1 alpha (PGC-1α) by suppressing nicotinamide adenine dinucleotide (NAD+)/Sirtuin 1 (SIRT1) signaling. The elevated SREBP1 then upregulated the expression of FASN, ACC1 and ELOVL6 in HCC cells, while PGC-1α/PPARα suppressed the expression of CPT1A and ACOX1.
Conclusions
Increased mitochondrial fission plays a crucial role in the reprogramming of lipid metabolism in HCC cells, which provides strong evidence for the use of this process as a drug target in the treatment of this malignancy.
Abbreviations
-
- ACC
-
- acetyl-CoA carboxylase
-
- ACOX1
-
- acyl-CoA oxidase 1
-
- ACSL4
-
- acyl-CoA synthetase long-chain family member 4
-
- CACT
-
- carnitine-acylcarnitine translocase
-
- chREBP
-
- carbohydrate-responsive element-binding protein
-
- CL
-
- cholesterol
-
- CPT1
-
- carnitine palmitoyl transferase 1
-
- CPT1A
-
- carnitine palmitoyl transferase 1A
-
- CPT2
-
- carnitine palmitoyl transferase 2
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- DRP1
-
- dynamin-related protein 1
-
- ELOVL6
-
- elongation of very long chain FA protein 6
-
- FA
-
- fatty acid
-
- FAO
-
- fatty acid oxidation
-
- FASN
-
- fatty acid synthase
-
- FBS
-
- fetal bovine serum
-
- FFA
-
- free fatty acid
-
- HCC
-
- hepatocellular carcinoma
-
- HFD
-
- high-fat diet
-
- HMG
-
- three-hydroxy-3-methylglutaryl
-
- HMGCR
-
- HMG-CoA reductase
-
- HMGCS1
-
- HMG-CoA synthase 1
-
- IHC
-
- immunohistochemistry
-
- LD
-
- lipid droplet
-
- MFN1
-
- downregulated mitofusin-1
-
- NAD
-
- nicotinamide adenine dinucleotide
-
- NASH
-
- non-alcoholic steatohepatitis
-
- PL
-
- phospholipid
-
- RT-PCR
-
- real-time PCR
-
- SCD1
-
- stearoyl-CoA desaturase 1
-
- SDS-PAGE
-
- sodium dodecyl sulphate-polyacrylamide gel electrophoresis
-
- SEM
-
- standard error of the mean
-
- shRNA
-
- short hairpin RNA
-
- SIRT1
-
- sirtuin 1
-
- SREBP
-
- sterol regulatory element-binding protein
-
- STR
-
- short tandem repeat
-
- TG
-
- triglyceride
1 BACKGROUND
Lipid metabolism reprogramming, an increasingly recognized hallmark of cancer, plays a critical role in cancer progression [1, 2]. Alterations in lipid metabolism in cancer cells are mainly characterized by high rates of de novo fatty acid (FA) synthesis and cholesterol biosynthesis, as well as altered FA degradation, which supports both proliferation and metastasis of cancer cells [3-5]. Aberrantly elevated expression of several key enzymes involved in de novo FA synthesis, such as acetyl coenzyme A (CoA) carboxylase (ACC), FA synthase (FASN), stearoyl-CoA desaturase 1 (SCD1), elongation of very long chain FA protein 6 (ELOVL6), three-hydroxy-3-methylglutaryl (HMG)-CoA synthase 1 (HMGCS1), and HMG-CoA reductase (HMGCR), has been observed in many different cancer types [6]. These lipogenic enzymes are under the transcriptional control of sterol regulatory element-binding proteins 1 and 2 (SREBP1 and SREBP2) [7]. Moreover, abnormal expression of enzymes involved in FA oxidation, such as peroxisome proliferator-activated receptor α (PPARα) target genes, carnitine palmitoyl transferase 1 (CPT1) and acyl-CoA oxidase 1 (ACOX1), has also been reported in several types of human cancers [8]. Sirtuin 1 (SIRT1) is a member of the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase that has been well demonstrated as a key regulator of lipid metabolism through the inhibition of crucial transcription factors or co-activators involved in de novo lipogenesis and FA β-oxidation, including SREBP1, PPARα, and PGC-1α [9]. Although lipid metabolism reprogramming has been widely acknowledged as an important characteristic of tumor cells, the underlying causes to SIRT1-mediated lipid metabolism remain incompletely understood.
Mitochondria are powerhouses in almost all eukaryotic cells. Their size and shape are formed dynamically by fission and fusion processes, which are tightly regulated to meet cellular metabolic demands [10, 11]. Recently, a series of studies have indicated that mitochondrial dynamic dysfunction is closely related to the development and progression of diverse pathological states, including cancer [12]. Our previous study has demonstrated that increased mitochondrial fission mediated by upregulated mitochondrial dynamin-related protein 1 (DRP1) and downregulated mitofusin-1 (MFN1) promoted tumor growth and metastasis of hepatocellular carcinoma (HCC) [13]. We have also demonstrated that suppression of mitochondrial fission reprogramed glucose metabolism from glycolysis to oxidative phosphorylation to promote tumor cell survival during energy stress [14], implying that mitochondrial fission plays a crucial role in the regulation of metabolism in cancer cells. However, the role of increased mitochondrial fission in the reprogramming of lipid metabolism in cancer cells remains largely unclear, especially in HCC.
In the present study, we aimed to systematically investigate the role and underlying molecular mechanisms of increased mitochondrial fission in the reprogramming of lipid metabolism in HCC cells. Additionally, the effects of reprogrammed lipid metabolism regulated by mitochondrial fission on tumor growth and metastasis were also explored.
2 MATERIALS AND METHODS
2.1 Patient tumor tissue collection and cell culture
HCC tissues were collected from 217 patients with HCC who underwent their initial surgery at Xijing Hospital affiliated to the Fourth Military Medical University (Xi'an, Shaanxi, China). Patients who received preoperative chemo- or radiotherapy were excluded from our study. This study was approved by the Research Ethics Committee of the hospital, and written informed consent was obtained from all the participants.
Human HCC cell lines (Huh-7, HLF, HLE, SNU-354, SNU-368, and SNU-739) were acquired from the Korean Cell Line Bank (Seoul, Republic of Korea) and the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan), and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) or Roswell Park Memorial Institute-1640 medium (RPMI-1640; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA) with 10% fetal bovine serum (FBS; Sangon Biotech, Shanghai, China) at 37°C in 5% CO2. All cell lines were validated using short tandem repeat (STR) profiling and mycoplasma detection.
2.2 Knockdown or overexpression of target genes
For knockdown or overexpression of DRP1 and MFN1, short hairpin RNA (shRNA) or forced expression vectors were constructed as described previously [13]. shRNA sequences used in this study were as follows: 5'-GCCATCGACTACATTCGCTTT-3' for shSREBP1; 5'-CUACUUCCUGAAAACAAC-3' for shDRP1; 5'-CCGTTATCTGAAGAGTTCCTGCAAGAAAT-3' for shPPARα. The shRNA containing specific sequences targeting the human SREBP1, DRP1 or PPARα mRNA sequence was cloned into the pSilenc- er™ 3.1-H1 puro vector (Ambion, Austin, TX, USA). A control shRNA was also cloned into the pSilenc- er™ 3.1-H1 puro vector, which was used as a silencing negative control. For overexpression, the coding sequences of DRP1, MFN1 and SREBP1 were amplified from cDNA derived from SMMC7721 cells using primers listed in Supplementary Table S1 and cloned into the pcDNA™3.1(+) vector (Invitrogen, Carlsbad, CA, USA). Transfection was performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions. Stable cell lines transfected with shRNAs or forced expression vectors were generated after selection with G418 (Sigma-Aldrich, St. Louis, MO, USA) and used for in vitro mechanistic investigations or in vivo studies.
Transient transfections of small interfering RNAs (siRNAs) of DRP1 and MFN1 were used to analyze the role of mitochondrial fission in lipid metabolism regulation in HCC cells in vitro. The sequences of siRNA for DRP1, and MFN1 are provided in Supplementary Table S2. The siRNAs were mixed with Lipofectamine 3000 reagent in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). After incubation at room temperature for 20 mins, the mixture was added to HCC cells. After 48 h incubation, the knockdown efficiency was tested by Western blotting.
2.3 Real-time PCR (RT-PCR)
Total RNA was extracted from HCC cell lines and tumor tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Then, mRNA was reversely transcribed into cDNA using the PrimeScript RT reagent kit (Takara, Kyoto, Japan). RT-PCR was performed using the SYBR-Green Master mix (Takara), in a real-time PCR instrument (Bio-Rad, Hercules, CA, USA) with the conditions of 95°C for 3 min, then 35 cycles of 95°C for 15 s and 58°C for 30 s. The relative expression of target genes was calculated using the 2−△△Ct method, and the housekeeping gene β-actin was used as an internal control. The primers used are listed in Supplementary Table S2.
2.4 Western blotting
Protein lysates were prepared with radioimmunoprecipitation assay buffer (RIPA) lysis buffer (Beyotime, Shanghai, China), and their concentrations were determined using the bicinchoninic acid assay (BCA) method. A total of 50 μg of protein was electrophoresed on 10% polyacrylamide gels for sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto methylcellulose membranes (Millipore, Billerica, MA, USA). The membranes were probed with the appropriate primary antibodies listed in Supplementary Table S3 and horseradish peroxidase-conjugated secondary antibodies (Maixin biotechologles, Fujian, Fuzhou, China). Detection was achieved using an enhanced chemiluminescence detection kit (Millipore, Billerica, MA, USA). For detection of the acetylation levels of SREBP1, PPARα and PGC-1α, Protein G Agarose and specified antibodies were incubated with the cell lysates overnight at 4°C. Then, the resins were washed and eluted. The bound proteins were separated by SDS-PAGE for Western blotting with acetylated lysine (Immunechem, Columbia, Canada).
2.5 Mitochondrial morphology analysis
The mitochondrial morphology of HCC cells was evaluated by confocal immunofluorescence microscopy (Olympus Corporation, Tokyo, Japan) using the fluorescent dye MitoTracker Green FM (Molecular Probes, Waltham, MA, USA) or transmission electron microscopy (FEI, cHillsboro, Oregon, USA) as described previously [14]. ImageJ software (NIH, Bethesda, MD, USA) was used for analysis of mitochondria length.
2.6 Free FA (FFA), triglyceride (TG) and phospholipid (PL) and cholesterol (CL) analyses
HCC cells were harvested using radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China). The levels of FA, TG, PL and CL were detected using EnzyChromTM FA, TG, PL and CL kits (Bioassay Systems, Hayward, CA, USA), respectively, according to the manufacturer's instructions. Briefly, chloroform/methanol (2:1) was added to cell lysis buffer followed by vortex for 30 sec and centrifuged 5 min at 14,000 rpm. Supernatant was removed, and the levels of FA, TG, PL and CL were then determined with the corresponding Kit. Measurements were normalized to cell numbers in different groups.
2.7 Neutral lipid detection assay
Cells were seeded onto dishes and fixed with 4% paraformaldehyde (Beyotime, Shanghai, China) for 20 min. After incubation in the fluorescent lipophilic dye BODIPY 493/503 solution (5 ng/mL, Thermo Fisher) for 1 h at 37°C in the dark, images were generated using an Olympus FV-1000 confocal microscope (Olympus Corporation, Tokyo, Japan). Results are expressed as relative fluorescence, which was quantified using the ImageJ software.
2.8 Oil red O staining
Oil red O staining was used to determine lipid droplet (LD) formation in HCC cells. Briefly, the HCC cells were plated in dishes and fixed with 4% paraformaldehyde for 20 min. Subsequently, the HCC cells were washed with PBS and stained with the Oil red O staining solution (Beyotime, Shanghai, China) for 20 min at room temperature. After washing with PBS, images were obtained with a light microscope (Olympus Corporation, Tokyo, Japan).
2.9 Assessment of fatty acid oxidation (FAO)
FAO and lipid content analyses in HCC cells were performed as previously described previously [15]. Briefly, cells were plated into 6-well plate at a density of 2 × 106 cells per well). Cells were then cultured in 600 μL of DMEM medium containing 1 μCi [9, 10(n)-3H] oleic acid (Amersham Pharmacia Biotech, Milano, Italy) for 12 hours. The aqueous phase containing 3H2O in the supernatant was extracted using chloroform/methanol (2:1 v/v). Then, scintillation solution was added and radioactivity was determined with a L6500 scintillation counter (Beckman Coulter, Brea, CA).
2.10 Immunohistochemistry (IHC) analysis
Paraffin-embedded HCC tumor tissue sections were deparaffinized with xylene, and rehydrated in ethanol. The tissues sections were then boiled in citrate buffer (pH 6.0; Sigma-Aldrich, St. Louis, MO, USA) for 15 min for antigen retrieval. After blocking with peroxidase solution (DAMAO, Tianjin, China) for 10 min at room temperature, the sections were treated with protein blocking solution (Maixin biotechologles, Fuzhou, Fujian, China) for 20 min at room temperature. Subsequently, the corresponding primary antibodies (Supplementary Table S3) were added and incubated overnight at 4°C, followed by detection with an IHC detection kit (Maixin biotechologles, Fuzhou, Fujian, China) according to the manufacturer's instructions. IHC staining results were viewed using a light Olympus microscope. Staining intensity was quantified independently by two pathologists in a double-blinded fashion.
2.11 Cell Proliferation (MTS) and colony formation assays
To assess cell viability, 103HCC cells were seeded into 96-well plates. After overnight culture, cell viability in each well was measured using a Cell Proliferation Kit (Promega, Madison, WI, USA) following the manufacturer's protocol. Briefly, cell culture medium was removed and cells were washed with phosphate buffer saline (PBS) buffer. Cells were then incubated with 200 μL of MTS reaction solution for 2 h. The absorbance at 490 nm was measured at each time point.
To assess colony formation ability, 1000 HCC cells were plated into 6-well plates. After culturing for 2 weeks, colonies were fixed with 4% paraformaldehyde, then stained with 0.5% crystal violet solution (Beyotime, Shanghai, China), and their numbers were counted using the ImageJ software.
2.12 Cell migration and invasion assay
To determine cell migration, a scratch wound healing assay was performed. A total of 2 × 105 HCC cells were plated into 6-well plates. After incubation for 24 h, wounds were made with a 200 μL-pipette tip at the bottom of the wells. Relative cell migration in each group was evaluated using a light Olympus microscope 48 h after wounding.
To determine cell invasion, 24-well chambers with Matrigel (Corning, Cambridge, MA, USA) at the dilution ratio of 1:6 (Matrigel: DMEM medium) were used. HCC cells were plated in upper chambers (Corning, Cambridge, MA, USA). After incubation for 48 h, the non-invaded cells above the insert membrane were scraped with a cotton swab. The invaded cells at the membranes were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution. Penetrated cells were photographed using a light Olympus microscope, and the cell number in each well was counted.
2.13 In vivo tumor growth and metastasis models
Six-week-old male nude mice (BALB/c) were purchased from the animal center of the Fourth Military Medical University and randomly divided into groups. The animals were maintained in temperature- and humidity-controlled specific pathogen-free condition on a 14-h dark/10-h light cycle and at 25°C. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee of Fourth Military Medical University in Xi'an, Shaanxi, China.
For evaluation of in vivo tumor growth, a total of 1 × 107 HCC cells in 500 μL DMEM medium were subcutaneously injected into the flank of the 6-week-old male BALB/c nude mice. Tumor volumes were measured every 5 days for 40 days. At the end of the experiment, the mice were euthanized by cervical dislocation, and the tumors were removed and weighed.
For evaluation of in vivo metastasis, 5 × 106 HCC cells were intravenously injected into the tail vein of 6-week-old male BALB/c nude mice. After 6 weeks, the mice were euthanized by cervical dislocation, and their lungs were harvested for hematoxylin and eosin staining (Baso, Wuhan, Changsha, China). The number of metastatic nodules was counted under a light Olympus microscope.
2.14 Usage of inhibitor and agonist
The SIRT1 inhibitor EX527, SIRT1 agonist resveratrol and PPARα agonist GW7647 were all purchased from Sigma-Aldrich (St. Louis, MO, USA). EX527, resveratrol, GW7647 were dissolved in a special solvent composed of 1% DMSO, and then added to the HCC cells medium at a concentration of 2 μmol/L (EX527), 20 μmol/L (resveratrol) and 2 μmol/L (GW7647) for 6 h.
2.15 Bioinformatic analysis based on TCGA data using KEGG pathway enrichment
The gene expression data from 374 tumors and 50 normal liver tissues were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Genes that are significantly associated with mitochondrial fission (indicated by expression ratio of DRP1 MFN1) were evaluated using R software. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for the annotation of mitochondrial fission-related genes.
2.16 Analysis of Fatty Acids by Gas chromatography–Time of Flight mass spectrometry (GC-TOF-MS)
Cells cultured in 6-well plate were subjected to three freeze–thaw cycles, and then 1.5 μg (13C2)-myristic acid in methanol containing (900 μL) of (13C2)- was added. After centrifugation, supernatant was collected and evaporated to dryness. The dried residue was then used for GC-TOF-MS analysis.
2.17 Statistical analysis
Each experiment was conducted three times, and the data are expressed as mean ± standard error of the mean (SEM). All analyses were performed using the SPSS software (version 17.0; SPSS, Chicago, IL, USA). For statistical comparisons between two or more groups, an unpaired two-tailed Student's t-test or one-way analysis of variance (ANOVA) with Tukey's post-hoc test was used, respectively. Differences were considered significant at P < 0.05.
3 RESULTS
3.1 Mitochondrial fission significantly increased lipid contents in HCC cells
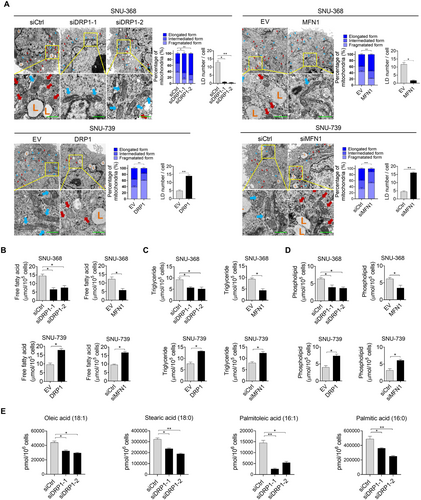
To investigate the role of increased mitochondrial fission in the regulation of aberrant lipid metabolism in HCC cells, bioinformatic analysis and experimental investigations were performed. Bioinformatic analysis based on TCGA data using KEGG pathway enrichment revealed that genes significantly associated with mitochondrial fission were notably involved in lipid metabolism (Supplementary Figure S1A), indicating that mitochondrial fission plays a crucial role in FA metabolic reprogramming in HCC cells. To select the most suitable cell line models for study, mitochondrial morphology was analyzed by staining with MitoTracker Green in a series of HCC cells lines. The results indicated that SNU-368 and SNU-739 cells have the most fragmented and elongated mitochondria, respectively. (Supplementary Figure S1B). Accordingly, we suppressed mitochondrial fission in SNU-368 cells by DRP1 knockdown or MFN1 overexpression. Meanwhile, we activated mitochondrial fission in SNU-739 cells by DRP1 overexpression or MFN1 knockdown. Successful knockdown or overexpression of DRP1 and MFN1 in SNU-368 or SNU-739 cells was tested by Western blotting analysis (Supplementary Figure S1C). The effect of mitochondrial fission on lipid content was then determined. Significantly decreased LDs were observed when mitochondrial fission was suppressed in SNU-368 cells, while accumulated LDs were observed when mitochondrial fission was activated in SNU-739 cells (Figure 1A and Supplementary Figure S1D). Similar results were also obtained by staining with the fluorescent lipophilic dye BODIPY 493/503, showing that mitochondrial fission elevated the intracellular content of neutral lipids in HCC cells (Supplementary Figure S1E). Consistently, decreased intracellular levels of FFA, TG, and PL were also observed when mitochondrial fission was suppressed, while the opposite effects on intracellular levels of FFA, TG, and PL were observed when mitochondrial fission was activated (Figure 1B-D). However, no significant change in the intracellular level of cholesterol was observed when mitochondrial fission was suppressed or activated (Supplementary Figure S1F). Moreover, metabolomic analysis by GC-TOF-MS indicated that mitochondrial fission also significantly increased the levels of FFA, including oleic acid (C18:1), stearic acid (C18:0), palmitoleate acid (C16:1), and palmitic acid (C16:0) (Figure 1E). Collectively, mitochondrial fission significantly increased the lipid content in human HCC cells.
3.2 Mitochondrial fission promoted de novo lipogenesis in HCC cells
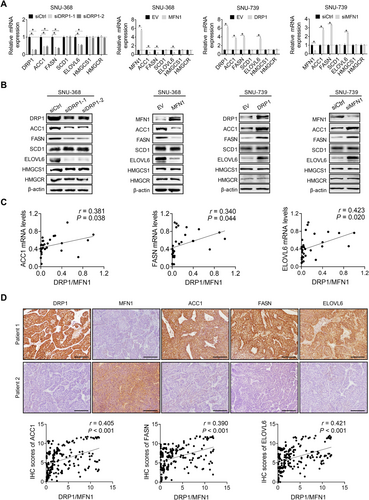
Increased lipid accumulation in HCC cells could be caused by elevated de novo FA synthesis and cholesterol biosynthesis. Thus, we assessed the role of mitochondrial fission in the regulation of de novo FA synthesis and cholesterol biosynthesis, both of which have been shown to be activated in many tumor types[6]. The expression of key enzymes involved in FA synthesis (FASN, ACC1, SCD1, and ELOVL6) and cholesterol biosynthesis (HMGCS1 and HMGCR) was detected by RT-PCR and Western blotting analyses. The data revealed that the suppression of mitochondrial fission significantly reduced the expression of FASN, ACC1, and ELOVL6 at both the mRNA and protein levels, while the expression of SCD1, HMGCS1, and HMGCR was unchanged (Figure 2A-B). In contrast, the activation of mitochondrial fission significantly increased the expression levels of FASN, ACC1, and ELOVL6 (Figure 2A-B), indicating that mitochondrial fission promoted de novo FA synthesis, although it had no effect on cholesterol biosynthesis in HCC cells. To provide further support, the mRNA levels of DRP1, MFN1, FASN, ACC1, and ELOVL6 were measured by RT-PCR analysis in tumor tissues from 30 patients with HCC. Scatter plot analysis indicated that mitochondrial fission (expression ratio of DRP1 to MFN1) was positively correlated with the mRNA levels of FASN, ACC1, and ELOVL6 (Figure 2C). Similar results were also obtained from IHC staining of DRP1, MFN1, FASN, ACC1, and ELOVL6 for their protein expression levels in tumor tissue sections from 217 HCC patients, showing significantly positive correlations between mitochondrial fission (expression ratio of DRP1 to MFN1) and the protein expression levels of FASN, ACC1, and ELOVL6 (Figure 2D).
3.3 Mitochondrial fission promoted de novo FA synthesis by upregulating SREBP1-mediated lipogenic gene expression
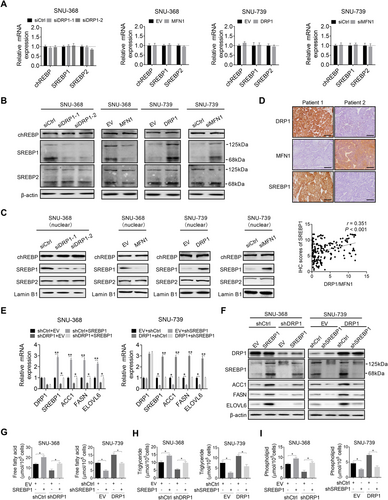
To understand the molecular mechanism underlying increased de novo FA synthesis, the changes in the expression levels of carbohydrate-responsive element-binding protein (chREBP) and SREBP1/2 [16], the crucial lipogenic transcriptional regulators, were determined when mitochondrial fission was suppressed or activated in HCC cells. The SREBP1 expression was significantly decreased only at the protein level in SNU-368 cells when mitochondrial fission was suppressed, while the chREBP and SREBP2 expression were unchanged at both the mRNA and protein levels (Figure 3A-B). In contrast, the activation of mitochondrial fission markedly increased the SREBP1 expression at the protein level in SNU-739 cells (Figure 3A-B). A similar expression pattern of SREBP1 in the nucleus was also observed (Figure 3C), indicating that the transcriptional activity of SREBP1 was induced by mitochondrial fission in HCC cells. Additionally, mitochondrial fission (expression ratio of DRP1 to MFN1) was also positively correlated with the expression of SREBP1 at the protein level in HCC tumor tissues (Figure 3D). Next, we explored whether the increased expression of SREBP1 contributed to the mitochondrial fission-induced upregulation of de novo FA synthesis. The results indicated that knockdown of SREBP1 markedly attenuated the promoting effect of increased mitochondrial fission on the expression of FASN, ACC1, and ELOVL6, as well as the contents of intracellular neutral lipids, FA, TG, and PL. In contrast, SREBP1 overexpression significantly reversed the downregulation of FASN, ACC1, and ELOVL6 (Figure 3E-F), as well as the intracellular levels of neutral lipids (Supplementary Figure S2). FA, TG, and PL were mediated by the suppression of mitochondrial fission (Figure 3G-I).
3.4 Mitochondrial fission suppressed FAO in HCC cells
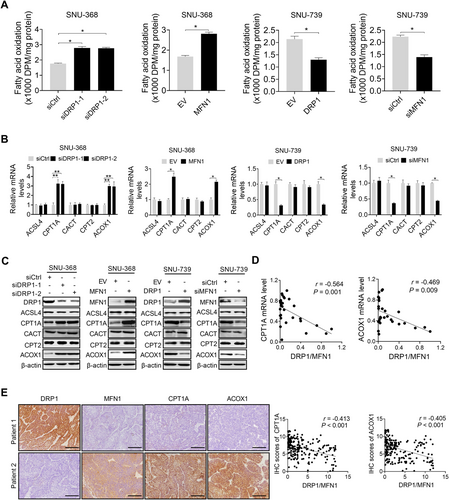
Given that decreased lipid catabolism may also contribute to the increased accumulation of lipids in HCC cells, we next investigated whether mitochondrial fission also regulates FAO in HCC cells. We observed that the suppression of mitochondrial fission significantly increased FAO in SNU-368 cells, as determined by the rate of 3H2O generation with 3H-labeled oleic acid as a tracer. Conversely, the activation of mitochondrial fission significantly inhibited FAO in SNU-739 cells (Figure 4A). The RT-PCR and Western blotting analyses for the expression of key regulators of FAO (acyl-CoA synthetase long-chain family member 4 (ACSL4), carnitine palmitoyl transferase 1A (CPT1A), carnitine-acylcarnitine translocase (CACT), carnitine palmitoyl transferase 2 (CPT2), and acyl-CoA oxidase 1 (ACOX1) demonstrated that the suppression of mitochondrial fission remarkably upregulated the CPT1A and ACOX1 expression in SNU-368 cells, while it had no significant effect on the ACSL4, CACT, and CPT2 expression (Figure 4B-C). In contrast, the activation of mitochondrial fission significantly downregulated CPT1A and ACOX1 expression in SNU-739 cells (Figure 4B-C). To provide further support, the CPT1A and ACOX1 expression levels were determined by RT-PCR and IHC analyses in tumor tissues from HCC patients. Our results indicated that mitochondrial fission (expression ratio of DRP1 to MFN1) was negatively correlated with the expression levels of CPT1A and ACOX1 at both the mRNA and protein levels (Figure 4D-E).
3.5 Mitochondrial fission suppressed FAO by downregulating peroxlsome proliferator-activated receptor (PPAR)-γ coactlvator-1α (PGC-1α)/PPARα signaling and its transcriptional targets
To understand the mechanisms underlying the suppression of FAO by mitochondrial fission in HCC cells, we analyzed the expression of critical hepatic lipid oxidation factor of PPARα and its co-activator PGC-1α in HCC cells with suppressed or activated mitochondrial fission. Suppression of mitochondrial fission had no significant effect on PPARα expression at either the mRNA or protein level, but significantly increased PGC-1α expression at the protein level (Figure 5A-B). In contrast, activation of mitochondrial fission significantly decreased PGC-1α expression at the protein level (Figure 5A-B), indicating that mitochondrial fission inhibits PGC-1α/PPARα signaling in HCC cells. As expected, a negative correlation existed between mitochondrial fission (expression ratio of DRP1 to MFN1) and PGC-1α expression at the protein level in the tumor tissues of patients with HCC (Figure 5C). Next, we investigated whether the reduced PGC-1α was involved in mitochondrial fission-downregulated CPT1A and ACOX1, both of which are transcriptional targets of PPARα and involved in the suppression of FAO in HCC cells. We found that the forced expression of PGC-1α markedly reversed the inhibitory effects of mitochondrial fission on the expression of CPT1A and ACOX1 and the rate of FAO in HCC cells (Figure 5D-F). Meanwhile, treatment with the PPARα agonist GW7647 phenocopied the reverse effect of forced expression of PGC-1α on mitochondrial fission-suppressed FAO in HCC cells, implying that mitochondrial fission suppresses FAO by downregulating PGC-1α/PPARα signaling (Figure 5G-I). Additionally, both PGC-1α overexpression and GW7647 treatment strongly attenuated the mitochondrial fission-increased intracellular contents of FA, TG, and PL (Figure 5J) and neutral lipids (Supplementary Figure S3A and S3B) in HCC cells.
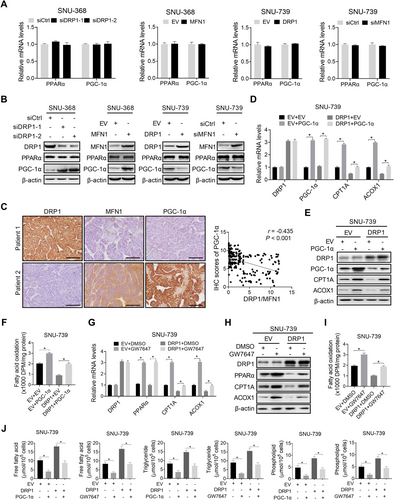
3.6 Mitochondrial fission mediated FA metabolic reprogramming though suppression of NAD+/SIRT1
Previously, mitochondrial fission has been reported to suppress oxidative phosphorylation and the production of NAD+, which is critical for the activity of SIRTs [17]. To better understand the mechanisms by which mitochondrial fission regulates lipid metabolism in HCC cells, we tested whether SIRT1 is involved in mitochondrial fission-mediated lipid metabolism reprogramming. The results revealed that the suppression of mitochondrial fission significantly increased NAD+ levels and SIRT1 activity in SNU-368 cells, while the activation of mitochondrial fission markedly decreased NAD+ levels and SIRT1 activity in SNU-739 cells (Figure 6A-B). Additionally, the suppression of mitochondrial fission clearly elevated the acetylation of SREBP1 and PGC-1α in SNU-368 cells, although it had no evident effect on the acetylation of PPARα (Figure 6C). In contrast, the activation of mitochondrial fission remarkably decreased the acetylation of SREBP1 and PGC-1α in SNU-739 cells (Figure 6C). As expected, the acetylation of SREBP1 and PGC-1α, the expression of their transcriptional targets (Figure 6D), the intracellular contents of neutral lipids (Supplementary Figure S4), FA, TG, and PL (Figure 6E-G), which are regulated by mitochondrial fission, were markedly reversed when SIRT1 was suppressed by treatment with EX-527 (SIRT1 inhibitor) or activated by treatment with resveratrol (SIRT1 agonist). Altogether, these results indicated that mitochondrial fission reprogramed FA metabolism though suppression of NAD+/SIRT1 in HCC cells.
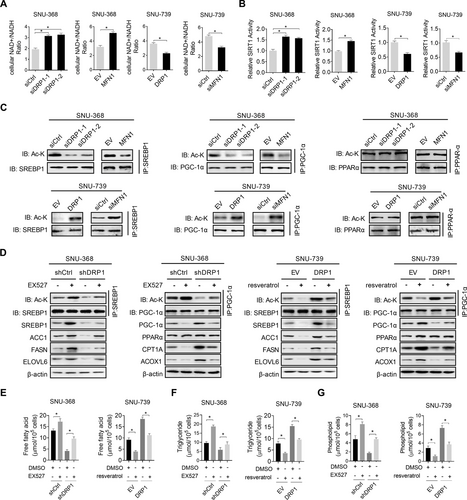
A recent study in fibroblasts has reported that NAD+-mediated activation of SIRT1 promoted DRP1 activation and thus mitochondrial fission [18]. Therefore, we tested whether a potential negative feedback loop exists between mitochondrial fission and NAD+/SIRT1 signaling in HCC cells. Unexpectedly, we did not observe any significant changes in DRP1 activation and mitochondrial morphology in HCC cells upon SIRT1 suppression or activation by treatment with EX-527 or resveratrol (Supplementary Figure S5), indicating that the relationship between NAD+/SIRT1 signaling and mitochondrial fission may be cell type-specific.
3.7 Mitochondrial fission promoted HCC growth and metastasis by mediating lipid metabolic reprogramming
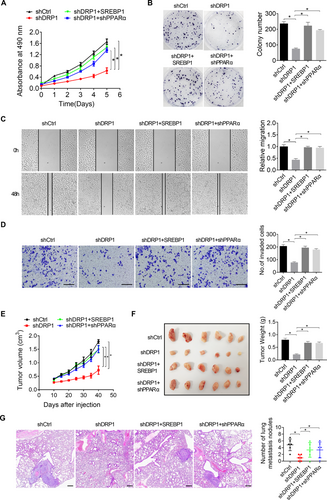
We further explored the contribution of lipid metabolism reprogrammed by increased mitochondrial fission to growth and metastasis of HCC cells. Our results indicated that elevated lipid content by forced SREBP1 expression or knockdown of PPARα markedly reversed the in vitro proliferation (Figure 7A), colony formation (Figure 7B), migration (Figure 7C), and invasion abilities (Figure 7D) of SNU-368 cells suppressed by mitochondrial fission inhibition. Additionally, mouse xenograft tumor growth and tail vein metastasis models were used to investigate the role of reprogrammed lipid metabolism in mitochondrial fission-promoted HCC growth and metastasis in vivo. The results showed that the suppressive effects of DRP1 knockdown on tumor growth and lung metastasis were markedly restored by forced SREBP1 expression or knockdown of PPARα (Figure 7E-G). Suppression of mitochondrial fission in dissected tumors was confirmed by transmission electron microscopy analysis, showing significantly increased mitochondrial length in DRP1 knockdown groups as compared with the control group (Supplementary Figure S6A). Additionally, we determined the production of NAD+, activity of SIRT1, lipid content, and the expression of lipid metabolic proteins including DRP1, SREBP1, ACC1, FASN, ELOVL6, PGC-1α, CPT1A, and ACOX1 in dissected tumors to confirm the in vitro findings from HCC cell lines. Markedly increased NAD+ levels (Supplementary Figure S6B), SIRT1 activity (Supplementary Figure S6C), and expression of lipid oxidative proteins PGC-1α, CPT1A, and ACOX1 (Supplementary Figure S6D), while decreased lipid content (Supplementary Figure S6E-F) and expression of lipogenic proteins SREBP1, ACC1, and FASN (Supplementary Figure S6D) were observed in the xenograft tumors with DRP1 knocking-down compared with those in the control group. The decreases in lipid content and changes in metabolic enzymes were all significantly reversed by overexpression of SREBP1 or knockdown of PPARα (Supplementary Figure S6A-S6F). Altogether, these findings suggest that lipid metabolism reprogramming plays a critical role in mitochondrial fission-promoted HCC growth and metastasis.
4 DISCUSSION
In the present study, we revealed that increased mitochondrial fission significantly enhanced de novo FA synthesis and suppressed FAO, which resulted in the accumulation of lipid content in HCC cells. However, a previous study has also reported that disordered mitochondrial dynamics mediated by the loss of MFN1 and MFN2 leads to impaired lipid synthesis in lung alveolar type 2 epithelial cells, which exacerbated bleomycin-induced lung fibrosis [19]. Inhibition of mitochondrial fission resulted in an increase in the FA content of LDs and a decrease in mitochondrial FAO. In addition, a recent study in using HeLa cells has reported that inhibition of mitochondrial fission increased the content of LDs and suppressed mitochondrial FAO [20]. Moreover, it has been shown that hepatocyte-specific deletion of mitochondrial fission factor in mice reduced hepatic triacylglycerol secretion, which promoted high-fat diet (HFD)-induced non-alcoholic steatohepatitis (NASH) phenotype [21]. These contradictions indicate a cell type-specific regulation of lipid metabolism by mitochondrial fission. Altogether, these results indicated that mitochondrial dynamic dysfunction plays a crucial role in the reprogramming of lipid metabolism and the progression of diverse human diseases, including cancer.
Normal cells depend on FA uptake, whereas tumor cells produce most of the FA from de novo synthesis to fulfill their need for rapid growth and proliferation [22]. Increased de novo FA synthesis and cholesterol biosynthesis have been reported in many types of human cancers, including HCC [2, 23]. Moreover, the upregulated expression of key enzymes involved in FA synthesis and cholesterol biosynthesis, such as FASN, ACC1, SCD1, ELOVL6, HMGCS1, and HMGCR, has also been observed in various human cancers [24]. In HCC, aberrant overexpression of lipogenic enzymes, such as FASN, ACC, SCD1, and HMGCR, has also been demonstrated. Consistently, our present study revealed that the increased mitochondrial fission by DRP1 overexpression or MFN1 knockdown promoted the de novo lipogenesis of HCC cells by upregulating key enzymes involved in FA synthesis (FASN, ACC1, SCD1, and ELOVL6), providing evidence of a molecular link between impaired mitochondrial dynamics and metabolic abnormalities.
SREBPs are key transcriptional regulators of lipogenesis and have three isoforms, SREBP1a, SREBP1c, and SREBP2. SREBP1 and SREBP2 play different roles in lipid synthesis [25]. SREBP1 transcriptionally activates genes involved in FA synthesis, whereas SREBP2 stimulates the transcription of genes involved in cholesterol biosynthesis. Overexpression of SREBP1 and SREBP2 has been demonstrated in several human cancer types [26-28]. Genetic or pharmacological inhibition of either SREBP1 or SREBP2 suppresses tumor cell growth and cell proliferation [27, 29]. In HCC, the expression of SREBP1 and SREBP2 has also been found to be gradually upregulated from non-neoplastic surrounding livers to HCC, which was closely associated with poor prognosis for patients with HCC [26]. Consistent with these findings, our results also indicated that increased mitochondrial fission significantly upregulated SREBP1 in HCC cells. It is well known that the activation of SREBPs by diminished cholesterol in ER promotes their translocation into the nucleus, where they transactivate various target lipogenesis genes by binding to sterol regulatory elements. Consistently, we also found that the expression of SREBP1 in the nucleus was activated by increased mitochondrial fission in HCC cells, which resulted in the upregulation of FA synthesis enzymes (FASN, ACC1, SCD1, and ELOVL6).
While accumulating studies have largely focused on the role of lipogenesis in the development and progression of cancer, the relevance of FAO to cancer has received less attention [30]. Since proliferating cancer cells require lipids to fulfill their need for rapid growth and proliferation, a reduction in FAO in cancer cells is expected. A previous study using HCC cells based on whole-genome microarray analysis has revealed the downregulation of FAO genes in HCC [31]. Additionally, it has also been demonstrated that suppression of FAO by hypoxia-inducible factor 1 plays a crucial role in HCC growth [32]. Consistently, we also found that mitochondrial fission suppressed FAO in HCC cells. The PPAR family, consisting of PPARα, PPARδ, and PAPR-γ, has been well established to play critical roles in hepatic lipid oxidation through transcriptional regulation of key enzymes involved in FAO [33]. Additionally, PPARα has been shown to play either a suppressive or a promotive role in cancer development depending on the tumor types [34]. In HCC cells, the suppression of PPARα has been reported to promote tumor progression [35]. Consistently, we also demonstrated that mitochondrial fission suppressed FAO in HCC cells by downregulating the expression of PPARα and its target genes CPT1A and ACOX1. We also demonstrated that mitochondrial fission-regulated suppression of FAO contributed to HCC growth and metastasis.
NAD+ is produced mainly by electron transporters in mitochondrial respiration [36]. Several previous studies have indicated that increased mitochondrial fission reduced mitochondrial respiratory chain function in both cell lines and mouse tissues, implying that mitochondrial fission may reduce NAD+ levels. Given that SIRT1 is known to be an NAD+-dependent protein deacetylase that plays important roles in lipid metabolism regulation, we explored the role of SIRT1 in mitochondrial fission-reprogrammed lipid metabolism. We found that mitochondrial fission suppressed SIRT1 activity by decreasing NAD+ production, which increased the acetylation and expression of SREBP1 and subsequently its transcriptional target lipogenic genes FASN, ACC1, SCD1, and ELOVL6. Consistently, a previous study has also demonstrated that SREBP1 was a direct target of SIRT1, and deacetylation of SREBP1 by SIRT1 inhibited SREBP1c expression and transactivation by decreasing its stability [37]. A recent study has reported that NAD+-mediated activation of SIRT1 promoted DRP1 activation and thus mitochondrial fission in fibroblasts [18]. However, our results showed that changes in SIRT1 activity by treatment with EX-527 or resveratrol had no significant effect on DRP1 activation and mitochondrial morphology in HCC cells, implying a cell type-specific regulation between NAD+/SIRT1 signaling and mitochondrial fission. In contrast to decreased SREBP1 expression by SIRT1-medicated acetylation, our results indicated that SIRT1-mediated acetylation increased the expression of PGC-1α. Consistent with our findings, a previous study has also demonstrated that SIRT1 deacetylated PGC-1α and increased its protein expression level [38]. These observations support the interpretation that SIRT1 plays diverse roles in the regulation of cellular lipid metabolism.
Previous studies by our group and others have demonstrated that increased mitochondrial fission plays an important role in tumor growth and metastasis [13, 39, 40]. Considering that lipids function not only as major building blocks but also as signaling molecules in cancer cells, we speculate that mitochondrial fission-reprogrammed lipid metabolism may contribute to the growth and metastasis of HCC cells. As expected, our results indicated that the decrease in lipid content by either forced expression of PPARα or knockdown of SREBP1 robustly attenuated mitochondrial fission-promoted growth and metastasis of HCC cells.
However, there are a few limitations in the present study. Firstly, the effects of increased mitochondrial fission on lipid metabolism were only measured in vitro HCC cell lines, the in vivo effects of increased on lipid metabolism in HCC mouse models were lacking. Secondly, although the major subject of this study is the effects of increased mitochondrial fission on lipid metabolism reprogramming, whether there exists a regulatory loop between mitochondrial fission and lipid metabolism reprogramming in HCC cells remain unclear. Therefore, additional experiments are needed in the future.
In conclusion, we demonstrated the crucial role of increased mitochondrial fission in the reprogramming of lipid metabolism in HCC cells, which provides strong evidence for the use of this process as a drug target in the treatment of this malignancy.
DECLARATIONS
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81772618), the Young Elite Scientist Sponsorship Program by CAST (2018QNRC001), and the State Key Laboratory of Cancer Biology Project (CBSKL2019ZZ26).
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS' CONTRIBUTIONS
LJB and XJL contributed to the study design, data collection, study analysis, manuscript writing, the funding to support the project and final approval of the manuscript submission. WD and YY conducted the mainly experiments, analyzed the data. HYR, ZZF and NL conducted the part of experiments in vitro and analyzed the data. YP and YT interpreted the patient samples and clinical data. All authors are responsible for the respective data and approved the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the ethics committee of Fourth Military Medical University (permit number: KY20173189-1). The tissue samples were obtained with written informed consent from each patient. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of Fourth Military Medical University (permit number: IACUC-20171207).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.




