ZNF582 hypermethylation promotes metastasis of nasopharyngeal carcinoma by regulating the transcription of adhesion molecules Nectin-3 and NRXN3
Abstract
Background
Epigenetic regulation plays an important role in the development and progression of nasopharyngeal carcinoma (NPC). However, the epigenetic mechanisms underlying NPC metastasis remains poorly understood. We aimed to find functional genes which regulate the metastasis of NPC and identify therapeutic targets for NPC treatment.
Methods
Bisulfite pyrosequencing was used to analyze zinc finger protein 582 (ZNF582) methylation in NPC tissues and cell lines. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blotting were used to determine the expression of ZNF582. In vitro and in vivo experiments were performed to evaluate the biological function of ZNF582 in NPC. ZNF582-targeting genes were identified by chromatin immunoprecipitation sequencing (ChIP-seq) and were confirmed by ChIP-qPCR and luciferase assay.
Results
ZNF582 promoter was hypermethylated in NPC, and both the mRNA and protein levels of ZNF582 were down-regulated in NPC tissues and cell lines. The restoration of ZNF582 inhibited NPC migration, invasion, and metastasis, while the knockdown of ZNF582 promoted NPC migration, invasion, and metastasis in vitro and in vivo. ZNF582 directly regulated the transcription and expression of adhesion molecules Nectin-3 and NRXN3. Both Nectin-3 and NRXN3 were identified as functional targets of ZNF582, and the restoration or abrogation of these genes reversed the tumor suppressor effect of ZNF582 in NPC metastasis.
Conclusions
ZNF582 acts as a tumor suppressor gene in NPC by regulating the transcription and expression of adhesion molecules Nectin-3 and NRXN3, which may provide novel therapeutic targets for NPC treatment.
Abbreviations
-
- NPC
-
- Nasopharyngeal carcinoma
-
- Co-IP
-
- Co-immunoprecipitation
-
- qRT-PCR
-
- Quantitative RT-PCR
-
- ZNF582
-
- Zinc Finger Protein 582
-
- DAC
-
- 5-aza-2'-deoxycytidine
1 BACKGROUND
Nasopharyngeal carcinoma (NPC) is a malignant head and neck tumor originating from the nasopharyngeal mucosal lining, and its distribution is geographically different [1-3]. When treated with intensity-modulated radiotherapy and chemotherapy, the local recurrence of NPC has been effectively controlled [1, 4, 5]. However, distant metastasis of NPC after treatment remains a key challenge [6-8]. Revealing the molecular mechanisms of NPC metastasis would help developing novel therapeutic strategies for NPC.
Our previous study has identified several hypermethylated and hypomethylated genes using the genome-wide methylated microarray chip analysis with 24 NPC tissues and 24 normal nasopharyngeal epithelial tissues (GSE52068) [9]. Based on microarray data, we found that the transcription factor zinc finger protein 582 (ZNF582), a member of the zinc finger protein family, was significantly hypermethylated in NPC tissues. To date, little is known about the function of ZNF582. Previous reports have revealed the hypermethylation of ZNF582 in cervical [10], esophageal [11], and oral cancers [12]. However, the function and mechanism of ZNF582 in these tumors, including NPC, remains completely unknown.
Cell adhesion molecules have been reported to participate in a variety of cellular functions, including morphogenesis [13], organogenesis [14, 15], and tumor progression [16, 17]. Nectin-3, which belongs to the nectin family, is a Ca2+-independent immunoglobulin (Ig)-like cell-cell adhesion molecule and is known to form adhesion junctions (AJs) by cooperating with E-cadherin [17-19]. Nectin-3 was shown to interact with afadin, which is an F-actin-binding protein, to form AJs. AJs mediate the formation and maintenance of tight junctions, which are part of the cell-cell adhesion process [20, 21]. AJs play key roles in gathering biologically active molecules, such as cell surface receptors, intracellular signaling molecules, and oncoproteins [22-24]. Previous studies have shown that nectins are involved in the adhesion, migration, and invasion of multiple types of cells [17]. Furthermore, the expression level of Nectin-3 has been reported to be associated with poor prognosis of lung adenocarcinoma [25] and pancreatic adenocarcinoma [26]. However, the role of Nectin-3 in NPC remains unknown. NRXN3 is a presynaptic adhesion molecule, which belongs to the neurexin genes (NRXN1, NRXN2, and NRXN3), and mainly regulates neurotransmitter release [27]. Recent studies have revealed that NRXN3 is also involved in the tumor progression of colorectal cancer [28], lung cancer [29], and glioblastoma [30]. However, the functional role of NRXN3 in NPC remains unknown.
In this study, we aimed to reveal how ZNF582 regulates NPC metastasis and figure out the relationship between ZNF582 and adhesion molecules in NPC.
2 MATERIALS AND METHODS
2.1 Clinical specimens and cell culture
In the present study, 27 normal nasopharynx tissue samples and 33 freshly frozen NPC biopsy tissue samples were obtained from the Sun Yat-sen University Cancer Center (Guangzhou, Guangdong, China). The present study was approved by the Institutional Ethical Review Board of Sun Yat-sen University Cancer Center, and informed consent was signed by all patients.
CNE2, SUNE1, HONE1, HNE1, 5-8F, and 6-10B NPC cell lines were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, ExCell Bio, Shanghai, China), and two human normal nasopharyngeal epithelial (NPE) cell lines (NP69 and N2-Tert) were maintained in keratinocyte serum-free medium (KSFM, Invitrogen) supplemented with bovine pituitary extract (BD Biosciences, Becton, NJ, USA). NP69, N2-Tert, and all NPC cell lines were generously provided by professor Mu-sheng Zeng (Sun Yat-sen University Cancer Center). HEK293T cells were grown in DMEM supplemented with 10% FBS and were obtained from American type culture collection (ATCC). All the cells were authenticated using shot-tandem repeat profiling, tested for mycoplasma contamination, and cultured for less than 2 months.
2.2 Constructs, antibodies, and reagents
ZNF582 or Nectin-3 were cloned into a Phage-puro-6tag vector via standard molecular methods. The Phage-puro-6tag vector was gifted by Dr. Xiaodong Zhang (Wuhan University, Wuhan, Hubei, China) ZNF582, Nectin-3, or NRXN3 shRNAs were cloned into the pLKO.1 vector (TranSheepBio, Shanghai, China). The Nectin-3 or NRXN3 promoter (the upstream 1,000 bp starting from the transcription starting site of Nectin-3 or NRXN3) was cloned into the pGL3-basic vector (Promega, Madison, WI, USA). The shRNA sequences are listed in Supplementary Table S1. Site-directed mutagenesis was performed with bridge-PCR and sequenced for confirmation. The anti-ZNF582 (10263, Sigma, St. Louis, MO, USA), anti-FLAG (F3165, Sigma), horseradish peroxide(HRP)-conjugated goat anti-mouse IgG (7076, Cell Signaling Technology, Boston, MA, USA), HRP-conjugated goat anti-rabbit IgG (7074, Cell Signaling Technology), anti-NRXN3 (DF9682, Affinity, Shanghai, China), anti-α-tubulin (11224-1-AP, Proteintech, Wuhan, Hubei, China), anti-Nectin-3 (11213-1-AP, Proteintech), 5-aza-2'-deoxycytidine (DAC, 189825, Sigma), Pierce™ Magnetic ChIP Kit (26157, Thermo, Waltham, MA, USA), Nano-Glo® Dual-Luciferase® Assay System (N1610, Promega), and puromycin (A1113802, Thermo) were purchased from the indicated manufacturers. The 5-week-old female BALB/c nude mice were obtained from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, Guangdong, China). The microarray data used in this study were downloaded from the GEO database using the GSE52068 and GSE62336 accession numbers.
2.3 DAC treatment
An amount of 1.5 × 105 NPC or NPE cells were seeded on 60 mm culture dishes (NEST Biotechnology, Wuxi, Jiangsu, China). After 24 h, the cells were treated with or without the methyltransferase inhibitor DAC (10 μmol/L) by replacing the drug every 24 h for 72 h. The cells were then collected for DNA and RNA extraction.
2.4 DNA extraction and bisulfite pyrosequencing
The genomic DNA was extracted from the NPC or NPE cells using the AllPrep RNA/DNA Mini kit (Qiagen, Dusseldorf, German) according to the manufacturer's instructions. The bisulfite transformation and purification of DNA (1-2 μg) were performed using the EpiTect Bisulfite Kit (Qiagen), following the manufacturer's instructions. The primers for polymerase chain reaction (PCR) and bisulfite pyrosequencing were designed by the PyroMark Assay Design Software 2.0 (Qiagen) and are listed in Supplementary Table S1. The PCR reactions were incubated at 95°C for 3 min, followed by 50 cycles of 15 s at 95°C, 20 s at 54°C and 30 s at 72°C, and a final 5 min hold at 72°C. The pyrophosphate sequencing reactions were conducted using the PyroMark Q96 ID system (Qiagen).
2.5 Real-time quantitative RT-PCR
RNA was extracted from NPE cells, NPC cells, and tissues using the TRIzol reagent (Invitrogen), and reverse transcription of the first-strand cDNA was performed using a reverse-transcription kit (Promega). The qRT-PCR assay was conducted on the Bio-Rad SFX (96 or 384) system with 2× SYBR Green mix (Life, Carlsbad, CA, USA). The data were normalized to the expression of GAPDH. The sequences of the primers are listed in Supplementary Table S1.
2.6 Lentivirus-mediated gene transfer
HEK293 cells were co-transfected with the indicated empty vector pLKO.1-shZNF582 or phage-6tag-ZNF582 and pSPAX2 and pMD2G (Addgene, Watertown, MA, USA). After 8-hour transfection, the culture medium was changed with fresh full medium. After an additional 8-hour transfection, the cell virus supernatants were harvested and used to infect SUNE1 or HONE1 cell lines. Then, 1 μg/mL puromycin was added to the medium to select the positive cells for 1-2 weeks. qRT-PCR and Western blotting assays were conducted to check the transfection efficiency.
2.7 Wound healing assay
SUNE1 or HONE1 cells were seeded in 6-well plates (NEST) to near confluence and then, incubated with serum-free medium for 24 h. Linear wounds were created using 200 μL tips in the cell monolayers, and the cells were washed twice with phosphate-buffered saline (PBS). The images were captured at 0 h and 24 h using a microscope (Leica, Wetzlar, Germany).
2.8 Migration and invasion assays
SUNE1, HONE1, NP69, or N2-Tert cells (5 × 104 or 1 × 105) were suspended in 200 μL of serum-free medium and seeded into upper Transwell chambers (8-μm pores, Corning, NY, USA) which were covered with (for invasion assay) or without matrigel (for migration assay, BD Biosciences). Then, the lower chambers were placed into 24-well plates (NEST) with a medium containing 10% FBS and incubated at 37°C. After 12 h (migration) or 24 h (invasion), these cells were fixed with methyl alcohol for 30 min and stained with crystal violet for 2 h. Migrated or invaded cells in 5 random visual fields were counted using a microscope (Leica).
2.9 Colony formation assay
SUNE1 (1000) or HONE1 cells (400) were seeded into a 6-well plate with full medium supplemented with 10% FBS, and cultured at 37°C for 1-2 weeks. Then, these cell colonies were fixed and stained with crystal violet. The images were captured using a microscope (Leica).
2.10 Cell adhesion assay
An empty 24-well plate with 10 μg/mL of fibronectin (Sigma) was incubated at 4°C overnight and washed thrice with PBS. Then, the plate was incubated with PBS containing 1% bovine serum albumin (BSA, Sigma) at 37°C for 1 h and washed thrice with PBS. Afterwards, SUNE1 or HONE1 cells (1 × 105) were seeded into the 24-well plate supplemented with serum-free medium and incubated at 37°C for 1 h. The non-adhesion cells that contained the medium were aspirated off. Then, these cells were gently washed with PBS, fixed with methyl alcohol, and stained with crystal violet. The images were captured using a microscope (Leica).
2.11 Chromatin immunoprecipitation assays (ChIP)
A Pierce™ Magnetic ChIP Kit (26157, Thermo) was used to perform the ChIP assay according to the manufacturer's protocol. Briefly, the cells were fixed with 1% formaldehyde and quenched with glycine. Then, these cells were washed twice with ice-cold PBS, detached by scraping in ice-cold PBS with 10 μL of Halt Cocktail, and harvested in ChIP lysis buffer for 10 min on ice. Afterwards, the lysate was centrifuged (9000 × g for 3 min) at 4°C, and the nuclei were resuspended for MNase Digestion in a 37°C water bath for 15 min. The reaction was stopped using the MNase Stop Solution and centrifuged (9000 × g for 5 min) at 4°C. Next, 1× IP dilution buffer (Thermo) containing protease/phosphatase inhibitors was added to resuspend the nuclei. Then, the suspension was sonicated on ice with several pulses and incubated for 20 s on ice between pulses. The suspensions were centrifuged (9000 × g for 5 min) at 4°C, and the supernatant containing the digested chromatin was harvested. Afterwards, the supernatant was incubated with normal rabbit IgG, or the anti-FLAG or anti-RNA polymerase II antibody at 4°C overnight. Then, ChIP-grade protein G magnetic beads (25 μL) were added into the lysate in the following morning, and these were incubated at 4°C for 2 h with mixing. The DNA was eluted using 1X IP elution buffer (150 μL) containing 5 mol/L NaCl (6 μL) and 20 mg/mL proteinase K (2 μL) through incubation at 65°C for 30 min with vigorous shaking. The eluted DNA was purified using a DNA Clean-UP column and DNA column wash buffer. The purified DNA was used to perform the qRT-PCR (ChIP-qRT-PCR) detection or sequencing (ChIP-seq). The sequences for the ChIP-qRT-PCR primers are listed in Supplementary Table S1.
2.12 Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
The above purified DNA was sequenced using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Then, the enriched genes (P < 0.05) were subjected to KEGG enrichment analysis by importing the list of genes into the DAVID website (https://david.ncifcrf.gov).
2.13 Reporter gene assay
SUNE1, HONE1, NP69, or N2-Tert cells (4 × 104) were seeded in 24-well plates for 12 h and co-transfected with plasmids encoding empty vector or FLAG-ZNF582 or shZNF582, along with the pGL3-Nectin-3 or NRXN3 reporter plasmid (100 ng) and a phRL-TK-Renella luciferase control vector (Addgene, 10 ng). After 24 h, the cells were lysed in 1 × passive lysis buffer (Promega), and the luciferase activities were determined using the Nano-Glo® Dual-Luciferase® Assay System (Promega). Afterwards, the firefly luciferase activity was normalized through the Renella luciferase activity, and the results were presented as the fold change relative to the activity in the empty vector-transfected cells.
2.14 Inguinal lymph node metastasis models
SUNE1 cells (3 × 105) stably encoding control shRNA (shControl) or shZNF582 were injected into the footpads of mice (n = 8 per group). These mice were euthanized after 35 days feeding, and the primary footpad tumors and linked inguinal lymph nodes were collected, fixed, paraffin-embedded, and sectioned. Hematoxylin-eosin (H&E) staining and immunohistochemistry (IHC) staining (anti-pan-cytokeratin antibody, Sigma) were conducted. The stained sections were captured using the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss, Jena, Germany). The popliteal lymph nodes with positive keratin IHC staining were considered as metastatic lymph nodes, and the metastatic popliteal lymph node ratio was defined as the proportion of metastatic lymph nodes among all resected popliteal lymph nodes.
2.15 In vivo xenograft tumor models
SUNE1 cells (1 × 106) stably encoding empty vector or ZNF582 were injected into the tail vein of mice. The survival of mice was recorded every day. After 8 weeks, all mice were euthanized, and the lung tissues were fixed, paraffin-embedded, and sectioned, then, H&E or IHC staining was conducted. The stained sections were analyzed using a Leica O8 microscope. The intensities of the staining were analyzed using the ViewPointBETA v1 software (Portland, OR, USA).
2.16 Immunohistochemistry assay
NPE and NPC paraffin tissue sections and xenograft mice tissue sections were used to perform the IHC assay. Briefly, these tissues were deparaffinized at 60°C for 30 min and rehydrated. Then, the endogenous peroxidase was blocked using 3% H2O2 for 30 min, and the tissues were maintained at high-temperature citrate for 2.5 min for antigen retrieval. Subsequently, the non-specific binding proteins were blocked with BSA, and the tissues were incubated with the indicated primary antibodies at 4°C overnight. The staining of the paraffin sections was performed as previously described [31]. The staining intensity score was defined as follows: 0, no staining; 1, weak, light yellow; 2, moderate, yellow-brown; and 3, strong, brown. The positive rate score was defined as follows: 1, < 10%; 2, 10%-35%; 3, 35%-70%; 4, > 70%. The total score of indicated proteins was calculated as staining intensity score × positive rate score.
2.17 Statistical analysis
Unpaired two-tailed Student's t-test and one-way or two-way ANOVA analysis with the SPSS 20 software were used to compare the statistical differences among groups. All data are presented as mean ± standard deviation (SD) and were extracted from no less than three independent experiments. A P value of less than 0.05 was considered significant. The animal survival curves were generated using the Kaplan-Meier method and analyzed by the log-rank analysis.
3 RESULTS
3.1 The methylation level of ZNF582 promoter in NPC
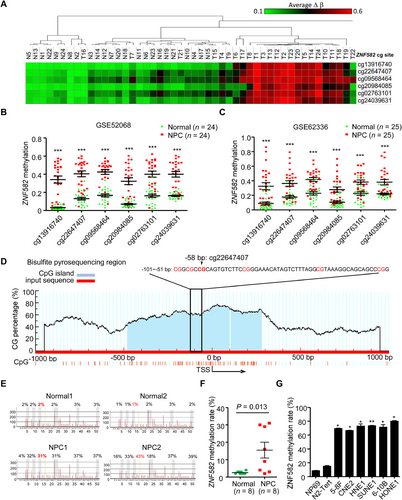
We analyzed the methylation status of ZNF582 in the genome-wide DNA methylation microarray dataset (GSE52068) [9], and found that ZNF582 promoter was frequently hypermethylated in NPC tissues (Figure 1A). Six significantly hypermethylated CpG sites were identified in the ZNF582 prompter region (Figure 1B). The results were confirmed by microarray analysis with the same six CpG sites conducted in the Hong Kong dataset (GSE62336, Figure 1C). To validate the dysregulated methylation status of ZNF582 in NPC, we performed the bisulfite pyrosequencing against cg22647407 (the most significantly hypermethylated CpG site of ZNF582 ranked by P value, Figure 1D). The methylation level of ZNF582 was significantly higher in NPC tissues than in normal tissues (Figure 1E-F). Consistently, the ZNF582 methylation was elevated in NPC cell lines (Figure 1G). Together, these data demonstrated that ZNF582 was hypermethylated in NPC.
3.2 Association between ZNF582 hypermethylation and its expression in NPC
We next determined whether the hypermethylation of ZNF582 would affect its expression in NPC. qRT-PCR and Western blotting assays were conducted to evaluate the expression of ZNF582 in cell lines and tissue samples. Both mRNA and protein levels of ZNF582 were significantly decreased in NPC cell lines and tissues (Figure 2A-D), indicating that ZNF582 was down-regulated in NPC.
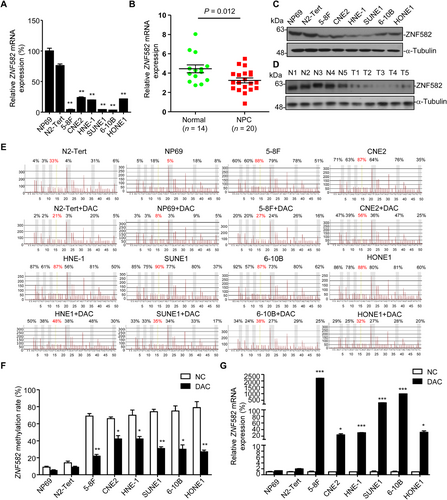
Next, we addressed whether the dysregulation of ZNF582 was caused by its promoter hypermethylation. After treatment with DAC, the ZNF582 methylation levels were significantly decreased in NPC cell lines (Figure 2E-F), whereas the mRNA levels of ZNF582 were substantially increased (Figure 2G), suggesting that ZNF582 was frequently down-regulated in NPC due to its promoter hypermethylation.
3.3 Association of ZNF582 with NPC cell migration and invasion in vitro
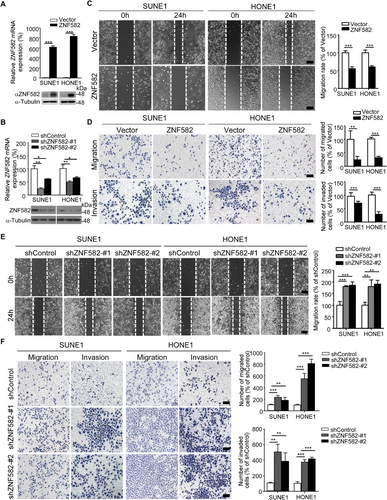
To determine the role of ZNF582 in the tumorigenesis or metastasis of NPC, colony formation and Transwell assays were performed with the NPC cell lines. The overexpression and two lentiviral shRNA constructs of ZNF582 were first generated, and the NPC cell lines with stable overexpression or knockdown of ZNF582 were established (Figure 3A-B). We found that the overexpression of ZNF582 significantly suppressed NPC cell migration and invasion (Figure 3C-D), while the knockdown of ZNF582 markedly increased the NPC cell migration and invasion (Figure 3E-F). However, neither the overexpression nor knockdown of ZNF582 had effect on NPC cell growth (data not shown). These results indicated that ZNF582 could inhibit NPC cell migration and invasion in vitro. To confirm these observations, we performed Transwell assays using normal nasopharyngeal epithelial cell lines (NP69 and N2-Tert) by knocking down ZNF582. Similar to the results in NPC cells, the knockdown of ZNF582 significantly increased the migration and invasion of NP69 and N2-Tert cells (Supplementary Figure S1). These results confirmed that ZNF582 inhibited the migration and invasion of both NPC and NPE cells.
3.4 Molecular mechanism of ZNF582 affecting NPC metastasis
Next, the mechanisms underlying the ZNF582-mediated suppressive effects in NPC were addressed. ChIP-seq against ZNF582 was performed to explore its downstream target genes, and a significant number of cell adhesion molecules were found to be highly enriched (Figure 4A). qRT-PCR confirmed that the adhesion molecule Nectin-3 was down-regulated (Figure 4B) and NRXN3 was up-regulated (Figure 4C) after ZNF582 overexpression in SUNE1 and HONE1 cells. Furthermore, after DAC treatment, the expression of ZNF582 was greatly increased in SUNE1 and HONE1 cells. Meanwhile, the expression of Nectin-3 was significantly inhibited and the expression of NRXN3 was greatly increased in SUNE1 and HONE1 cells (Supplementary Figure S2A), suggesting that ZNF582 could regulate the gene expression of Nectin-3 and NRXN3 at the mRNA level. To further validate these findings, we investigated the expression levels of Nectin-3 and NRXN3 in NP69 and N2-Tert cells. Compared with those in NP69 and N2-Tert cells, both the mRNA and protein levels of Nectin-3 were increased and NRXN3 were decreased in SUNE1 and HOEN1 cells (Supplementary Figure S2B-D). In addition, the knockdown of ZNF582 in NP69 or N2-Tert cells promoted the expression of Nectin-3 and inhibited the expression of NRXN3 (Supplementary Figure S2E-F). These results confirmed that ZNF582 could regulate the expression of Nectin-3 and NRXN3 on both mRNA and protein levels.
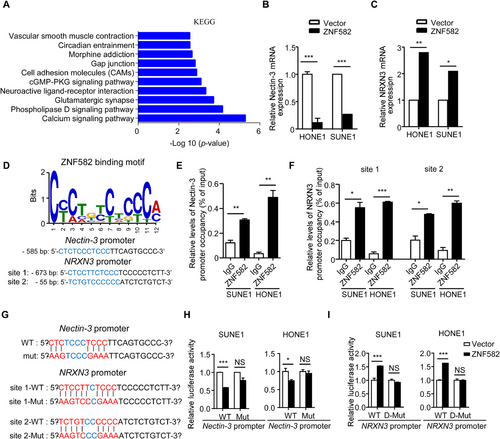
To assess how ZNF582 regulates the expression of Nectin-3 and NRXN3, ChIP-qPCR was performed. The binding motif of ZNF582 was predicted by previous CHIP-seq analysis, and one ZNF582-binding site in the Nectin-3 promoter and two ZNF582-binding sites in the NRXN3 promoter were identified (Figure 4D). The results revealed that ZNF582 could directly bind to the promoter regions of Nectin-3 and NRXN3 in NPC (Figure 4E-F) and NPE cells (Supplementary Figure S2G-H). To further confirm that ZNF582 drives the transcription of Nectin-3 and NRXN3, the upstream 1,000 bp (starting from the transcription starting site of Nectin-3 or NRXN3) was cloned to the pGL3 basic luciferase vector. Meanwhile, the constructs of Nectin-3 or NRXN3 promoter with mutations on the ZNF582-binding sites were made, and luciferase reporter assays were performed (Figure 4G). Consistent with previous qRT-PCR data in SUNE1 and HONE1 cells, ZNF582 was found to potently inhibit the luciferase activity of the wild-type Nectin-3 promoter, but not the mutant Nectin-3 promoter (Figure 4H). Meanwhile, ZNF582 significantly increased the luciferase activity of the wild-type NRXN3 promoter, but not the mutant NRXN3 promoter (Figure 4I). Furthermore, the knockdown of ZNF582 in NP69 and N2-Tert cells demonstrated opposite effects (Supplementary Figure S2I-J). Together, these data suggested that ZNF582 could directly bind to the promoter of Nectin-3 or/and NRXN3 and drove the transcriptional inhibition or activation of these genes.
3.5 Association of Nectin-3 with NPC cell adhesion, migration, and invasion in vitro
We analyzed the expression of Nectin-3 in NPE and NPC tissues archived in the GSE12452 dataset and found that Nectin-3 was significantly overexpressed in NPC tissue samples (Figure 5A) and was inversely correlated with ZNF582 expression (R = -0.310, P = 0.049) (Figure 5B), suggesting that Nectin-3 could be an oncogene in NPC. To identify the exact role of Nectin-3 in NPC, the adhesion, migration, and invasion abilities of NPC cells with the overexpression or knockdown of Nectin-3 were detected. The overexpression of Nectin-3 promoted the cell adhesion, migration, and invasion of NPC cells (Figure 5C-D), while the knockdown of Nectin-3 inhibited these abilities (Figure 5E). These results indicated that Nectin-3 acts as an oncogene in NPC.
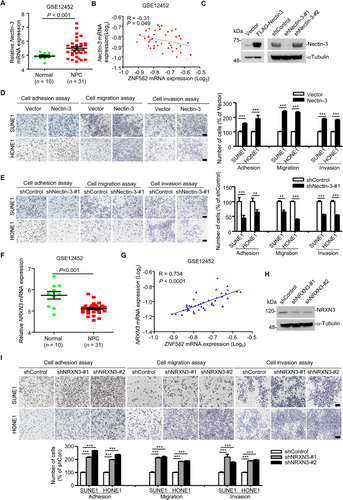
3.6 Association of NRXN3 with NPC cell adhesion, migration, and invasion in vitro
NRXN3 expression was lower in NPC tissues than in normal tissues (Figure 5F) and was positively correlated with ZNF582 expression in NPC tissues (GSE12452) (Figure 5G; R = 0.734, P < 0.001). We hypothesized that NRXN3 could function as a tumor suppressor gene in NPC. Cell adhesion, migration, and invasion assays were performed to identify the role of NRXN3 in NPC. As expected, the knockdown of NRXN3 increased the adhesion, migration, and invasion abilities of NPC cells (Figure 5H-I), indicating that NRXN3 acted as a tumor suppressor in NPC.
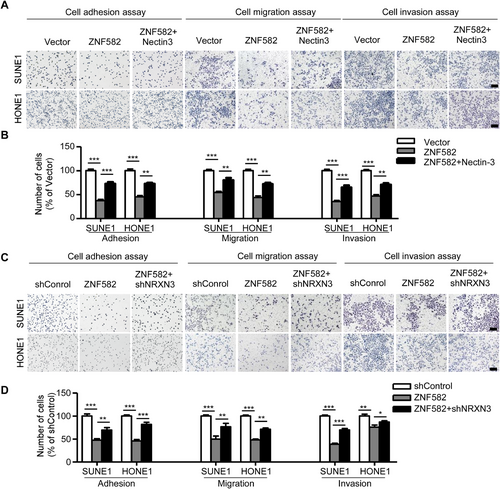
3.7 Both Nectin-3 and NRXN3 are functional targets of ZNF582 in NPC
We confirmed that ZNF582 induced the down-regulation of Nectin-3 by inhibiting its transcription. Hence, we determined the effects of Nectin-3 and NRXN3 on the tumor repressor function of ZNF582 in vitro. The overexpression of Nectin-3 (Figure 6A-B) and knockdown of NRXN3 (Figure 6C-D) reversed the suppressive effect of ZNF582 on the adhesion, migration, and invasion of NPC cells that stably overexpressed ZNF582, suggesting that both Nectin-3 and NRXN3 were functional targets of ZNF582 in NPC.
3.8 ZNF582 inhibits NPC cell invasion and metastasis in vivo
To validate the functions of ZNF582 in NPC in vivo, a popliteal lymph node metastasis model was constructed by transplanting SUNE1 cells stably expressing shZNF582 or shControl into the footpads of nude mice. After 35 days, the primary tumors and popliteal lymph nodes were obtained for analysis (Figure 7A). H&E staining revealed that the primary tumors in the ZNF582 knockdown group had a more aggressive phenotype with the invasion of tumor cells towards the skin and muscle, when compared to tumors in the shControl group (Figure 7B). Furthermore, the volume of the popliteal lymph nodes and the number of pan-cytokeratin-positive tumor cells were greater in the ZNF582 knockdown group than in the shControl group (Figure 7C-D). Strikingly, the metastatic popliteal lymph node ratio was remarkably higher in the ZNF582 knockdown group than in the shControl group (Figure 7E; P < 0.05).
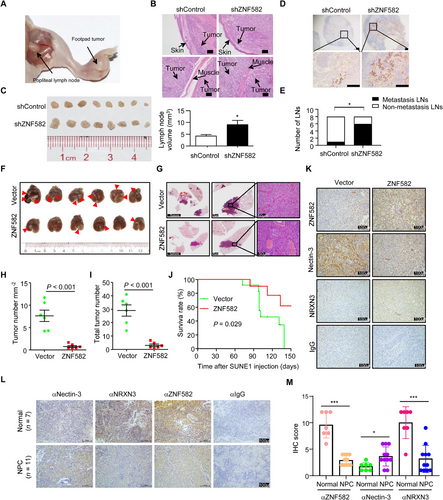
To further investigate the effect of ZNF582 on NPC metastasis in vivo, lung metastasis models were employed. Mice injected with SUNE1 that stably restored ZNF582 exhibited significantly reduced metastatic tumor colonies and tumor density in the lungs (Figure 7F-I) and longer survival (Figure 7J) when compared to mice injected with control cells, suggesting that ZNF582 could suppress NPC cell metastasis in vivo.
To further determine whether ZNF582 regulates the expression of Nectin-3 and NRXN3 in vivo, IHC staining was performed to detect the expression of these proteins in lung metastasis. Consistent with the present in vitro findings, the expression of Nectin-3 was significantly decreased and the expression of NRXN3 was significantly increased in the ZNF582 overexpression group, when compared with those in the vector group (Figure 7K). Moreover, the same results were obtained for the expression levels of Nectin-3 and NRXN3 in NPE and NPC tissue samples (Figure 7L-M). These results indicated that the up-regulation of ZNF582 reduced the expression of Nectin-3 and increased the expression of NRXN3 in vivo.
4 DISCUSSION
In this study, we identified that ZNF582 was hypermethylated in NPC, which resulted in its low expression. Functional studies showed that ZNF582 could inhibit the migration and invasion of NPC cells in vitro and in vivo. The downstream genes of ZNF582 (Nectin-3 and NRXN3) were found by ChIP-seq analysis. ZNF582 could directly bind to Nectin-3 and NRXN3 and regulate their transcription and expression. The restoration of Nectin-3 or abolition of NRXN3 in NPC cells that stably overexpressed ZNF582 could reverse the inhibitory effect of ZNF582 on NPC.
ZNF582 was first reported to be hypermethylated and may serve as a biomarker for cervical cancer [32, 33]. Emerging data suggested that ZNF582 was also hypermethylated in esophageal squamous cell carcinoma [11, 34] and oral cancer [12, 35]. In the present study, we also found that ZNF582 was hypermethylated in NPC and ZNF582 expression was significantly decreased in NPC tissues as compared with that in normal nasopharyngeal tissues. Moreover, we demonstrated that ZNF582 functions as a suppressor gene in NPC. However, the mechanism of ZNF582 in regulating cancer progression remains unknown. Zinc finger proteins is a large family of transcriptional factors which activate or repress gene expression by binding to gene promoters [36]. Previous studies have revealed that ZNF382, ZNF750, and ZNF671, which also belong to the zinc finger protein family, were involved in the regulation of gene transcription in NPC progression [37-39]. For instance, ZNF382 could interact with heterochromatin protein 1 and may be involved in heterochromatin formation and silencing [37]. ZNF750 binds to the promoter of fibroblast growth factor 14 (FGF14) to regulate its transcription and expression [39]. Therefore, we performed ChIP-Seq to identify the target genes of ZNF582. The results revealed that ZNF582 directly targets adhesion molecules Nectin-3 and NRXN3, thereby regulating its gene transcription and mRNA expression. Our findings suggest that ZNF582 functions as a transcription factor.
Nectins are Ca+-independent immunoglobulin-like cell adhesion molecules, including Nectin-1, Nectin-2, Nectin-3, and Nectin-4 [40]. They participate in multiple cellular processes, especially in regulating cell proliferation, migration, invasion, and tumor initiation [19, 41]. Loss of Nectin-3 expression is associated with aggressive phenotypes of the pancreatic neuroendocrine tumors [42]. Nectin-3 up-regulates matrix metallopeptidase 2(MMP2) and MMP9 and is a new biomarker in ovarian cancer [43]. What's more, Nectin-3 interacts with Necl-5 to promote the proliferation and invasion of colon adenocarcinoma cell line [44]. Accumulating evidence shows that Nectin-3 plays a vital role in tumor progression [45-47]. We found that ZNF582 directly bound to the promoter of Nectin-3 and inhibited its transcription and expression (Figure 4), indicating a new regulation mechanism of Nectin-3 expression. Besides, we demonstrated that Nectin-3 functioned as an oncogene in NPC through in vitro functional studies (Figure 5), and this may partially explain the antitumor effect of ZNF582 in NPC.
In this study, we also found another target of ZNF582, NRXN3, which is also a cell adhesion molecule. NRXN3 was first found to play a critical role in Alzheimer disease, as it is a presynaptic adhesion molecule that specifies neuron synapses and regulates neurotransmitter release [48-50]. However, NRXN3 was also found to play an important role in tumor progression, including colorectal cancer [28, 51], lung cancer [29], and glioblastoma [30, 52]. Our study found that NRXN3 expression was lower in NPC than in normal nasopharyngeal tissues, and knockdown of NRXN3 promoted NPC migration and invasion in vitro (Figure 5).
In the present study, in vitro functional studies demonstrated that Nectin-3 functioned as an oncogene, while NRXN3 functioned as a suppressor gene in NPC. Although both Nectin-3 and NRXN3 are cell adhesion molecules, the functions of them in NPC are completely opposite. We assume that ZNF582 regulates the two molecules by two independent processes, which needs to be proved by subsequent experiments. For further study, we will continue to explore the specific regulatory mechanisms of ZNF582 on Nectin-3 and NRXN3, and to elucidate the molecular mechanisms of Nectin-3 and NRXN3 underlying their functions in NPC. For instance, Nectin-3 has been reported to associate with F-actin through afadin, which is an F-actin-binding protein, and forms AJs [17, 53]. Therefore, ZNF582 may inhibit NPC cell adhesion by regulating the Nectin-3-afadin-F-actin association. Further studies are needed to deepen our understanding of the roles of ZNF582 in regulating adhesion molecules during NPC progression.
5 CONCLUSIONS
Hypermethylation of ZNF582 promoted NPC metastasis in vitro and in vivo. Furthermore, adhesion molecules Nectin-3 and NRXN3 function as the downstream targets of ZNF582 to regulate NPC metastasis. These findings provide new insights into the molecular mechanisms that govern NPC metastasis, which may further provide novel therapeutic strategies for NPC treatment.
ACKNOWLEDGEMENTS
Not applicable.
AUTHORS' CONTRIBUTIONS
N.L., J.M. and Y.Z. designed the research. Y.Z., X.H., K.L. and Y.L. conducted the experiments. Y.Z., Y.L., S.H., P.Z., J.L., Q.L., Y.L. and Y.C. acquired and analyzed the data. N.L. and J.M. provided the reagents. Y. Z., N.L. and J.M. wrote the manuscript. Y.Z. and Y.C. revised the manuscript.
COMPETING INTERESTS
The authors declare that no competing interests exist.
AVAILABILITY OF DATA AND MATERIALS
The microarray data used in the study was downloaded from the GEO database using accession numbers GSE52068 and GSE62336. The ZNF582, Nectin-3 and NRXN3 expression data in NPC were obtained from the GSE12452 dataset. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (http://www.researchdata.org.cn), with the approval RDD number as RDDB2020000818.
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Ethical Review Board of the Sun Yat-sen University Cancer Center, and all patients provided a signed informed consent. All animal studies were approved by the Institutional Animal Care and Use Ethics Committee of Sun Yat-sen University Cancer Center.
FUNDING
This study was supported by grants from the National Natural Science Foundation of China (81902962), the China Postdoctoral Science Foundation (2019M653224), the Planned Science and Technology Project of Guangdong Province (2019B020230002), the Natural Science Foundation of Guangdong Province (2017A030312003), the Health and Medical Collaborative Innovation Project of Guangzhou City, China (201803040003), the Innovation Team Development Plan of the Ministry of Education (IRT_17R110), and the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, B14035).




