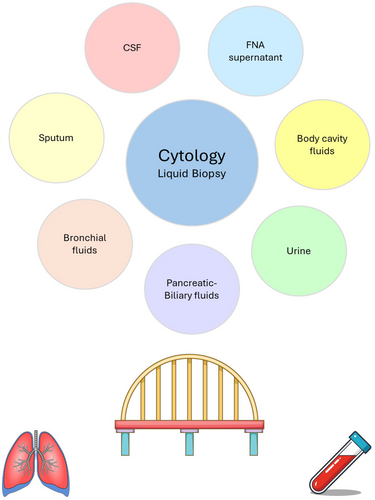
The Bridge: Supernatant Derived From Cytological Sample Preparations
Corresponding Author
Sinchita Roy-Chowdhuri
Division of Pathology and Laboratory Medicine, Department of Pathology, MD Anderson Cancer Center, The University of Texas, Houston, Texas, USA
Correspondence:
Sinchita Roy-Chowdhuri ([email protected])
Search for more papers by this authorCorresponding Author
Sinchita Roy-Chowdhuri
Division of Pathology and Laboratory Medicine, Department of Pathology, MD Anderson Cancer Center, The University of Texas, Houston, Texas, USA
Correspondence:
Sinchita Roy-Chowdhuri ([email protected])
Search for more papers by this authorABSTRACT
The scope and extent of molecular cytopathology in the era of precision medicine has been expanding in recent years. The versatility of cytology specimen preparations has provided ample opportunity for the cytopathology community to evolve, innovate and ‘do more with less’ using limited amounts of tissue. More recently, cytology-derived supernatant liquid biopsy samples have been identified as a substantial source of high-quality genomic material that can be interrogated for genotyping for therapeutic decision-making, as well as other roles in cancer screening for early-stage disease, longitudinal monitoring for therapeutic response and disease prognostication. These novel substrates, including supernatants from body fluids such as urine, pleural effusion, ascitic fluid, cerebrospinal fluid, as well as fine-needle aspiration (FNA) specimens, serve as a bridge between tissue-based testing and conventional liquid biopsy testing from the patient's plasma. Cytologically derived liquid biopsy samples can only be used in situations where the tissue sample is inadequate for genotyping, or when plasma-based liquid biopsy fails to identify an oncogenic driver alteration, but they can be used as a stand-alone complementary specimen source that can provide reliable genomic information for therapeutic decisions. This review aims to highlight some of the advances in the field and the clinical applications of the cytology-derived supernatant specimen.
Graphical Abstract
Cytology-derived supernatant specimens provide a substantial source of high-quality genomic material that can be used for genotyping to guide therapeutic decision-making, as well as for other roles in cancer screening, longitudinal monitoring and disease prognostication. These novel substrates, such as supernatants from cerebrospinal fluid, sputum, urine, pleural effusion, ascitic fluid, as well as fine-needle aspiration (FNA) specimens, serve as a bridge between tissue-based testing and conventional plasma-based liquid biopsy. Cytologically derived liquid biopsy samples can not only be used in situations where the tissue sample is inadequate for genotyping, or when plasma-based liquid biopsy fails to identify an oncogenic driver alteration, but they can be used as a stand-alone complementary specimen source that can provide reliable genomic information for therapeutic decisions.
Conflicts of Interest
The author declares no conflicts of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1E. Vigliar, U. Malapelle, C. de Luca, C. Bellevicine, and G. Troncone, “Challenges and Opportunities of Next-Generation Sequencing: A Cytopathologist's Perspective,” Cytopathology 26 (2015): 271–283.
- 2N. Rekhtman and S. Roy-Chowdhuri, “Cytology Specimens: A Goldmine for Molecular Testing,” Archives of Pathology & Laboratory Medicine 140 (2016): 1189–1190, https://doi.org/10.5858/arpa.2016-0379-ED.
- 3G. da Cunha Santos, “Standardizing Preanalytical Variables for Molecular Cytopathology,” Cancer Cytopathology 121 (2013): 341–343, https://doi.org/10.1002/cncy.21290.
- 4G. da Cunha Santos and M. A. Saieg, “Preanalytic Specimen Triage: Smears, Cell Blocks, Cytospin Preparations, Transport Media, and Cytobanking,” Cancer Cytopathology 125 (2017): 455–464, https://doi.org/10.1002/cncy.21850.
- 5C. Bellevicine, U. Malapelle, E. Vigliar, P. Pisapia, G. Vita, and G. Troncone, “How to Prepare Cytological Samples for Molecular Testing,” Journal of Clinical Pathology 70 (2017): 819–826, https://doi.org/10.1136/jclinpath-2017-204561.
- 6P. Pisapia, F. Pepe, R. Sgariglia, et al., “Next Generation Sequencing in Cytology,” Cytopathology 32 (2021): 588–595, https://doi.org/10.1111/cyt.12974.
- 7M. H. Roh, “The Utilization of Cytologic Fine-Needle Aspirates of Lung Cancer for Molecular Diagnostic Testing,” Journal of Pathology and Translational Medicine 49 (2015): 300–309, https://doi.org/10.4132/jptm.2015.06.16.
- 8I. Chebib and V. Y. Jo, “Round Cell Sarcoma With CIC-DUX4 Gene Fusion: Discussion of the Distinctive Cytomorphologic, Immunohistochemical, and Molecular Features in the Differential Diagnosis of Round Cell Tumors,” Cancer Cytopathology 124 (2016): 350–361, https://doi.org/10.1002/cncy.21685.
- 9C. M. Parseghian, A. L. Tam, J. Yao, et al., “Assessment of Reported Trial Characteristics, Rate of Publication, and Inclusion of Mandatory Biopsies of Research Biopsies in Clinical Trials in Oncology,” JAMA Oncology 5 (2019): 402–405, https://doi.org/10.1001/jamaoncol.2018.4640.
- 10M. Salto-Tellez, “Diagnostic Molecular Cytopathology - a Further Decade of Progress,” Cytopathology 26 (2015): 269–270, https://doi.org/10.1111/cyt.12276.
- 11F. C. Schmitt, “Demystifying Molecular Cytopathology,” International Journal of Surgical Pathology 18 (2010): 213S–215S, https://doi.org/10.1177/1066896910370887.
- 12P. A. Vander Laan, “Molecular Markers: Implications for Cytopathology and Specimen Collection,” Cancer Cytopathology 123 (2015): 454–460, https://doi.org/10.1002/cncy.21560.
- 13E. Vigliar, A. Iaccarino, M. Sciortino, et al., “Molecular Predictive Testing in Precision Oncology: The Italian Experience,” Cancer Cytopathology 128 (2020): 622–628, https://doi.org/10.1002/cncy.22290.
- 14S. K. Tian, J. K. Killian, N. Rekhtman, et al., “Optimizing Workflows and Processing of Cytologic Samples for Comprehensive Analysis by Next-Generation Sequencing: Memorial Sloan Kettering Cancer Center Experience,” Archives of Pathology & Laboratory Medicine 140 (2016): 1200–1205, https://doi.org/10.5858/arpa.2016-0108-RA.
- 15D. H. Hwang, E. P. Garcia, M. D. Ducar, E. S. Cibas, and L. M. Sholl, “Next-Generation Sequencing of Cytologic Preparations: An Analysis of Quality Metrics,” Cancer 125 (2017): 786–794, https://doi.org/10.1002/cncy.21897.
- 16S. Roy-Chowdhuri, H. Chen, R. R. Singh, et al., “Concurrent Fine Needle Aspirations and Core Needle Biopsies: A Comparative Study of Substrates for Next-Generation Sequencing in Solid Organ Malignancies,” Modern Pathology 30 (2017): 499–508, https://doi.org/10.1038/modpathol.2016.228.
- 17S. Roy-Chowdhuri, R. S. Goswami, H. Chen, et al., “Factors Affecting the Success of Next-Generation Sequencing in Cytology Specimens,” Cancer Cytopathology 123 (2015): 659–668, https://doi.org/10.1002/cncy.21597.
- 18S. Roy-Chowdhuri and J. Stewart, “Preanalytic Variables in Cytology: Lessons Learned From Next-Generation Sequencing,” Archives of Pathology & Laboratory Medicine 140 (2016): 1191–1199, https://doi.org/10.5858/arpa.2016-0117-RA.
- 19L. Durin, A. Pradines, C. Basset, et al., “Liquid Biopsy of Non-Plasma Body Fluids in Non-Small Cell Lung Cancer: Look Closer to the Tumor!,” Cells 9 (2020): 2486, https://doi.org/10.3390/cells9112486.
- 20P. Pisapia, F. Pepe, A. Iaccarino, et al., “Next Generation Sequencing in Cytopathology: Focus on Non-Small Cell Lung Cancer,” Frontiers in Medicine 8 (2021): 633923, https://doi.org/10.3389/fmed.2021.633923.
- 21P. A. VanderLaan and S. Roy-Chowdhuri, “Current and Future Trends in Non-Small Cell Lung Cancer Biomarker Testing: The American Experience,” Cancer Cytopathology 128 (2020): 629–636, https://doi.org/10.1002/cncy.22313.
- 22U. Malapelle, P. Pisapia, A. Addeo, et al., “Liquid Biopsy From Research to Clinical Practice: Focus on Non-Small Cell Lung Cancer,” Expert Review of Molecular Diagnostics 21 (2021): 1165–1178, https://doi.org/10.1080/14737159.2021.1985468.
- 23A. E. Brown, K. S. Lim, G. Corpus, M. T. Hustek, T. A. N. Tran, and C. C. Chang, “Detection of BRAF Mutation in the Cytocentrifugation Supernatant Fluid From Fine-Needle Aspiration of Thyroid Lesions May Enhance the Diagnostic Yield,” CytoJournal 14 (2017): 4, https://doi.org/10.4103/1742-6413.200935.
- 24G. Deftereos, S. D. Finkelstein, S. A. Jackson, et al., “The Value of Mutational Profiling of the Cytocentrifugation Supernatant Fluid From Fine-Needle Aspiration of Pancreatic Solid Mass Lesions,” Modern Pathology 27 (2014): 594–601, https://doi.org/10.1038/modpathol.2013.147.
- 25S. D. Finkelstein, M. Bibbo, T. E. Kowalski, et al., “Mutational Analysis of Cytocentrifugation Supernatant Fluid From Pancreatic Solid Mass Lesions,” Diagnostic Cytopathology 42 (2014): 719–725, https://doi.org/10.1002/dc.23048.
- 26N. Guibert, H. Tsukada, D. H. Hwang, et al., “Liquid Biopsy of Fine-Needle Aspiration Supernatant for Lung Cancer Genotyping,” Lung Cancer 122 (2018): 72–75, https://doi.org/10.1016/j.lungcan.2018.05.024.
- 27Z. Guo, Z. Xie, H. Shi, et al., “Malignant Pleural Effusion Supernatant Is an Alternative Liquid Biopsy Specimen for Comprehensive Mutational Profiling,” Thorac Cancer 10 (2019): 823–831, https://doi.org/10.1111/1759-7714.13006.
- 28B. Hannigan, W. Ye, M. Mehrotra, et al., “Liquid Biopsy Assay for Lung Carcinoma Using Centrifuged Supernatants From Fine-Needle Aspiration Specimens,” Annals of Oncology 30, no. 6 (2019): 963–969, https://doi.org/10.1093/annonc/mdz102.
- 29K. Hummelink, M. Muller, T. C. Linders, et al., “Cell-Free DNA in the Supernatant of Pleural Effusion Can Be Used to Detect Driver and Resistance Mutations, and Can Guide Tyrosine Kinase Inhibitor Treatment Decisions,” ERJ Open Research 5 (2019): 2019, https://doi.org/10.1183/23120541.00016-2019.
- 30A. Kawahara, H. Abe, K. Murata, et al., “Screening System for Epidermal Growth Factor Receptor Mutation Detection in Cytology Cell-Free DNA of Cerebrospinal Fluid Based on Assured Sample Quality,” Cytopathology 30 (2019): 144–149, https://doi.org/10.1111/cyt.12660.
- 31S. Roy-Chowdhuri, M. Mehrotra, A. M. Bolivar, et al., “Salvaging the Supernatant: Next Generation Cytopathology for Solid Tumor Mutation Profiling,” Modern Pathology 31 (2018): 1036–1045, https://doi.org/10.1038/s41379-018-0006-x.
- 32L. Tong, N. Ding, X. Tong, et al., “Tumor-Derived DNA From Pleural Effusion Supernatant as a Promising Alternative to Tumor Tissue in Genomic Profiling of Advanced Lung Cancer,” Theranostics 9 (2019): 5532–5541, https://doi.org/10.7150/thno.34070.
- 33W. Ye, B. Hannigan, S. Zalles, et al., “Centrifuged Supernatants From FNA Provide a Liquid Biopsy Option for Clinical Next-Generation Sequencing of Thyroid Nodules,” Cancer Cytopathology 127 (2019): 146–160, https://doi.org/10.1002/cncy.22098.
- 34L. Y. Ballester, I. C. Glitza Oliva, D. Y. Douse, et al., “Evaluating Circulating Tumor DNA From the Cerebrospinal Fluid of Patients With Melanoma and Leptomeningeal Disease,” Journal of Neuropathology and Experimental Neurology 77 (2018): 628–635, https://doi.org/10.1093/jnen/nly046.
- 35M. Shah, T. Takayasu, S. Zorofchian Moghadamtousi, et al., “Evaluation of the Oncomine Pan-Cancer Cell-Free Assay for Analyzing Circulating Tumor DNA in the Cerebrospinal Fluid in Patients With Central Nervous System Malignancies,” Journal of Molecular Diagnostics 23 (2021): 171–180, https://doi.org/10.1016/j.jmoldx.2020.10.013.
- 36P. S. Chauhan, K. Chen, R. K. Babbra, et al., “Urine Tumor DNA Detection of Minimal Residual Disease in Muscle-Invasive Bladder Cancer Treated With Curative-Intent Radical Cystectomy: A Cohort Study,” PLoS Medicine 18 (2021): e1003732, https://doi.org/10.1371/journal.pmed.1003732.
- 37I. J. Russo, Y. Ju, N. S. Gordon, et al., “Toward Personalised Liquid Biopsies for Urothelial Carcinoma: Characterisation of ddPCR and Urinary cfDNA for the Detection of the TERT 228 G>A/T Mutation,” Bladder Cancer 4 (2018): 41–48, https://doi.org/10.3233/BLC-170152.
- 38U. Satyal, A. Srivastava, and P. H. Abbosh, “Urine Biopsy-Liquid Gold for Molecular Detection and Surveillance of Bladder Cancer,” Frontiers in Oncology 9 (2019): 1266, https://doi.org/10.3389/fonc.2019.01266.
- 39R. Zhang, J. Zang, F. Xie, et al., “Urinary Molecular Pathology for Patients With Newly Diagnosed Urothelial Bladder Cancer,” Journal of Urology 206 (2021): 873–884, https://doi.org/10.1097/JU.0000000000001878.
- 40Z. Wu, Z. Yang, C. S. Li, et al., “Differences in the Genomic Profiles of Cell-Free DNA Between Plasma, Sputum, Urine, and Tumor Tissue in Advanced NSCLC,” Cancer Medicine 8 (2019): 910–919, https://doi.org/10.1002/cam4.1935.
- 41H. Gokozan, A. Harbhajanka, P. Bomeisl, C. W. Michael, and N. Sadri, “Use of Cytology Centrifuged Supernatants Improves Cost and Turnaround Time for Targeted Next Generation Sequencing,” Diagnostic Cytopathology 48 (2020): 1167–1172, https://doi.org/10.1002/dc.24548.
- 42N. Malhotra, S. A. Jackson, L. L. Freed, et al., “The Added Value of Using Mutational Profiling in Addition to Cytology in Diagnosing Aggressive Pancreaticobiliary Disease: Review of Clinical Cases at a Single Center,” BMC Gastroenterology 14 (2014): 135, https://doi.org/10.1186/1471-230X-14-135.
- 43E. Faber, H. Grosu, S. Sabir, et al., “Adequacy of Small Biopsy and Cytology Specimens for Comprehensive Genomic Profiling of Patients With Non-Small-Cell Lung Cancer to Determine Eligibility for Immune Checkpoint Inhibitor and Targeted Therapy,” Journal of Clinical Pathology 75, no. 9 (2021): 612–619, https://doi.org/10.1136/jclinpath-2021-207597.
- 44N. B. Leighl, R. D. Page, V. M. Raymond, et al., “Clinical Utility of Comprehensive Cell-Free DNA Analysis to Identify Genomic Biomarkers in Patients With Newly Diagnosed Metastatic Non-Small Cell Lung Cancer,” Clinical Cancer Research 25 (2019): 4691–4700, https://doi.org/10.1158/1078-0432.CCR-19-0624.
- 45M. H. Maher, D. Y. Duose, I. I. Wistuba, R. Luthra, S. Arjuna, and S. Roy-Chowdhuri, “A Rapid Turnaround Time Workflow for a Cytological Liquid Biopsy Assay Using FNA Supernatant Specimens,” Cancer Cytopathology 133 (2025): e22925, https://doi.org/10.1002/cncy.22925.
- 46C. Abbosh, N. J. Birkbak, and C. Swanton, “Early Stage NSCLC - Challenges to Implementing ctDNA-Based Screening and MRD Detection,” Nature Reviews. Clinical Oncology 15 (2018): 577–586, https://doi.org/10.1038/s41571-018-0058-3.
- 47A. Kawahara, C. Fukumitsu, T. Taira, et al., “Epidermal Growth Factor Receptor Mutation Status in Cell-Free DNA Supernatant of Bronchial Washings and Brushings,” Cancer Cytopathology 123 (2015): 620–628, https://doi.org/10.1002/cncy.21583.
- 48J. S. Ryu, J. H. Lim, M. K. Lee, et al., “Feasibility of Bronchial Washing Fluid-Based Approach to Early-Stage Lung Cancer Diagnosis,” Oncologist 24 (2019): e603–e606, https://doi.org/10.1634/theoncologist.2019-0147.
- 49C. K. Liam, Y. S. Liam, and C. K. Wong, “Extracellular Vesicle-Based EGFR Genotyping in Bronchoalveolar Lavage Fluid,” Translation Lung Cancer Research 9 (2020): 168–171, https://doi.org/10.21037/tlcr.2020.03.06.
- 50J. Y. Hur, J. S. Lee, I. A. Kim, et al., “Extracellular Vesicle-Based EGFR Genotyping in Bronchoalveolar Lavage Fluid From Treatment-Naive Non-Small Cell Lung Cancer Patients,” Translational Lung Cancer Research 8 (2019): 1051–1060, https://doi.org/10.21037/tlcr.2019.12.16.
- 51J. E. Kim, J. S. Eom, W. Y. Kim, et al., “Diagnostic Value of microRNAs Derived From Exosomes in Bronchoalveolar Lavage Fluid of Early-Stage Lung Adenocarcinoma: A Pilot Study,” Thorac Cancer 9 (2018): 911–915, https://doi.org/10.1111/1759-7714.12756.
- 52D. Dietrich, C. Kneip, O. Raji, et al., “Performance Evaluation of the DNA Methylation Biomarker SHOX2 for the Aid in Diagnosis of Lung Cancer Based on the Analysis of Bronchial Aspirates,” International Journal of Oncology 40 (2012): 825–832, https://doi.org/10.3892/ijo.2011.1264.
- 53B. Schmidt, V. Liebenberg, D. Dietrich, et al., “SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer Based on Bronchial Aspirates,” BMC Cancer 10 (2010): 600, https://doi.org/10.1186/1471-2407-10-600.
- 54A. J. Hubers, D. A. M. Heideman, S. Duin, et al., “DNA Hypermethylation Analysis in Sputum of Asymptomatic Subjects at Risk for Lung Cancer Participating in the NELSON Trial: Argument for Maximum Screening Interval of 2 Years,” Journal of Clinical Pathology 70 (2017): 250–254, https://doi.org/10.1136/jclinpath-2016-203734.
- 55A. J. Hubers, D. A. M. Heideman, S. A. Burgers, et al., “DNA Hypermethylation Analysis in Sputum for the Diagnosis of Lung Cancer: Training Validation Set Approach,” British Journal of Cancer 112 (2015): 1105–1113, https://doi.org/10.1038/bjc.2014.636.
- 56J. Liao, P. Dhilipkannah, and F. Jiang, “Improving CT Scan for Lung Cancer Diagnosis With an Integromic Signature,” Journal of Biological Methods 11 (2024): e99010023.
- 57D. Lissa and A. I. Robles, “Sputum-Based DNA Methylation Biomarkers to Guide Lung Cancer Screening Decisions,” Journal of Thoracic Disease 9 (2017): 4308–4310.
- 58A. Hulbert, I. Jusue-Torres, A. Stark, et al., “Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum,” Clinical Cancer Research 23 (2017): 1998–2005, https://doi.org/10.1158/1078-0432.CCR-16-1371.
- 59Z. Song, W. Wang, M. Li, J. Liu, and Y. Zhang, “Cytological-Negative Pleural Effusion Can Be an Alternative Liquid Biopsy Media for Detection of EGFR Mutation in NSCLC Patients,” Lung Cancer 136 (2019): 23–29, https://doi.org/10.1016/j.lungcan.2019.08.004.
- 60D. Liu, Y. Lu, Z. Hu, et al., “Malignant Pleural Effusion Supernatants Are Substitutes for Metastatic Pleural Tumor Tissues in EGFR Mutation Test in Patients With Advanced Lung Adenocarcinoma,” PLoS One 9 (2014): e89946, https://doi.org/10.1371/journal.pone.0089946.
- 61J. Lin, Y. Gu, R. du, M. Deng, Y. Lu, and Y. Ding, “Detection of EGFR Mutation in Supernatant, Cell Pellets of Pleural Effusion and Tumor Tissues From Non-Small Cell Lung Cancer Patients by High Resolution Melting Analysis and Sequencing,” International Journal of Clinical and Experimental Pathology 7 (2014): 8813–8822.
- 62A. Mokanszki, E. S. Bádon, A. Mónus, L. Tóth, N. Bittner, and G. Méhes, “Cell-Free DNA From Pleural Effusion Samples: Is It Right for Molecular Testing in Lung Adenocarcinoma?,” Pathology Oncology Research 27 (2021): 613071, https://doi.org/10.3389/pore.2021.613071.
- 63J. S. Lee, J. Y. Hur, I. A. Kim, et al., “Liquid Biopsy Using the Supernatant of a Pleural Effusion for EGFR Genotyping in Pulmonary Adenocarcinoma Patients: A Comparison Between Cell-Free DNA and Extracellular Vesicle-Derived DNA,” BMC Cancer 18 (2018): 1236, https://doi.org/10.1186/s12885-018-5138-3.
- 64S. Villatoro, C. Mayo-de-las-Casas, N. Jordana-Ariza, et al., “Prospective Detection of Mutations in Cerebrospinal Fluid, Pleural Effusion, and Ascites of Advanced Cancer Patients to Guide Treatment Decisions,” Molecular Oncology 13 (2019): 2633–2645, https://doi.org/10.1002/1878-0261.12574.
- 65P. Zhang, X. Wu, M. Tang, X. Nie, and L. Li, “Detection of EGFR Gene Mutation Status From Pleural Effusions and Other Body Fluid Specimens in Patients With Lung Adenocarcinoma,” Thoracic Cancer 10 (2019): 2218–2224, https://doi.org/10.1111/1759-7714.13201.
- 66L. De Mattos-Arruda, R. Mayor, C. K. Y. Ng, et al., “Cerebrospinal Fluid-Derived Circulating Tumour DNA Better Represents the Genomic Alterations of Brain Tumours Than Plasma,” Nature Communications 6 (2015): 8839, https://doi.org/10.1038/ncomms9839.
- 67J. Seoane, L. De Mattos-Arruda, E. Le Rhun, A. Bardelli, and M. Weller, “Cerebrospinal Fluid Cell-Free Tumour DNA as a Liquid Biopsy for Primary Brain Tumours and Central Nervous System Metastases,” Annals of Oncology 30 (2019): 211–218.
- 68B. Y. Jiang, Y. S. Li, W. B. Guo, et al., “Detection of Driver and Resistance Mutations in Leptomeningeal Metastases of NSCLC by Next-Generation Sequencing of Cerebrospinal Fluid Circulating Tumor Cells,” Clinical Cancer Research 23 (2017): 5480–5488, https://doi.org/10.1158/1078-0432.CCR-17-0047.
- 69Y. S. Li, B. Y. Jiang, J. J. Yang, et al., “Unique Genetic Profiles From Cerebrospinal Fluid Cell-Free DNA in Leptomeningeal Metastases of EGFR-Mutant Non-Small-Cell Lung Cancer: A New Medium of Liquid Biopsy,” Annals of Oncology 29 (2018): 945–952, https://doi.org/10.1093/annonc/mdy009.
- 70S. Ying, H. Ke, Y. Ding, et al., “Unique Genomic Profiles Obtained From Cerebrospinal Fluid Cell-Free DNA of Non-Small Cell Lung Cancer Patients With Leptomeningeal Metastases,” Cancer Biology & Therapy 20 (2019): 562–570, https://doi.org/10.1080/15384047.2018.1538614.
- 71C. Ma, X. Yang, W. Xing, H. Yu, T. Si, and Z. Guo, “Detection of Circulating Tumor DNA From Non-Small Cell Lung Cancer Brain Metastasis in Cerebrospinal Fluid Samples,” Thorac Cancer 11 (2020): 588–593, https://doi.org/10.1111/1759-7714.13300.
- 72A. M. Miller, R. H. Shah, E. I. Pentsova, et al., “Tracking Tumour Evolution in Glioma Through Liquid Biopsies of Cerebrospinal Fluid,” Nature 565 (2019): 654–658, https://doi.org/10.1038/s41586-019-0882-3.
- 73A. Di Meo, J. Bartlett, Y. Cheng, M. D. Pasic, and G. M. Yousef, “Liquid Biopsy: A Step Forward Towards Precision Medicine in Urologic Malignancies,” Molecular Cancer 16 (2017): 80, https://doi.org/10.1186/s12943-017-0644-5.
- 74H. Husain, V. O. Melnikova, K. Kosco, et al., “Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine,” Clinical Cancer Research 23 (2017): 4716–4723, https://doi.org/10.1158/1078-0432.CCR-17-0454.
- 75F. S. Togneri, D. G. Ward, J. M. Foster, et al., “Genomic Complexity of Urothelial Bladder Cancer Revealed in Urinary cfDNA,” European Journal of Human Genetics 24 (2016): 1167–1174, https://doi.org/10.1038/ejhg.2015.281.
- 76B. Liu, J. Ricarte Filho, A. Mallisetty, et al., “Detection of Promoter DNA Methylation in Urine and Plasma Aids the Detection of Non-Small Cell Lung Cancer,” Clinical Cancer Research 26 (2020): 4339–4348, https://doi.org/10.1158/1078-0432.CCR-19-2896.
- 77S. Roy-Chowdhuri, “Molecular Testing of Residual Cytology Samples: Rethink, Reclaim, Repurpose,” Cancer Cytopathology 127 (2019): 15–17, https://doi.org/10.1002/cncy.22076.
- 78S. Roy-Chowdhuri, “Tumor-Derived Cell-Free DNA in Body Cavity Effusion Supernatants: A Promising Alternative for Genomic Profiling,” Cancer Cytopathology 128 (2020): 14–16, https://doi.org/10.1002/cncy.22206.