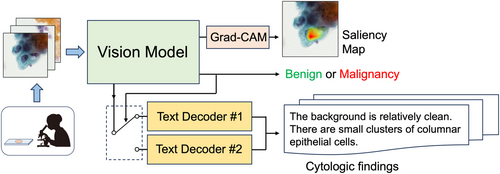
Automated Description Generation of Cytologic Findings for Lung Cytological Images Using a Pretrained Vision Model and Dual Text Decoders: Preliminary Study
Funding: This work was supported in part by a Grant-in-Aid for Scientific Research (No. 23K07117), MEXT, Japan.
ABSTRACT
Objective
Cytology plays a crucial role in lung cancer diagnosis. Pulmonary cytology involves cell morphological characterisation in the specimen and reporting the corresponding findings, which are extremely burdensome tasks. In this study, we propose a technique to generate cytologic findings from for cytologic images to assist in the reporting of pulmonary cytology.
Methods
For this study, 801 patch images were retrieved using cytology specimens collected from 206 patients; the findings were assigned to each image as a dataset for generating cytologic findings. The proposed method consists of a vision model and dual text decoders. In the former, a convolutional neural network (CNN) is used to classify a given image as benign or malignant, and the features related to the image are extracted from the intermediate layer. Independent text decoders for benign and malignant cells are prepared for text generation, and the text decoder switches according to the CNN classification results. The text decoder is configured using a transformer that uses the features obtained from the CNN for generating findings.
Results
The sensitivity and specificity were 100% and 96.4%, respectively, for automated benign and malignant case classification, and the saliency map indicated characteristic benign and malignant areas. The grammar and style of the generated texts were confirmed correct, achieving a BLEU-4 score of 0.828, reflecting high degree of agreement with the gold standard, outperforming existing LLM-based image-captioning methods and single-text-decoder ablation model.
Conclusion
Experimental results indicate that the proposed method is useful for pulmonary cytology classification and generation of cytologic findings.
Graphical Abstract
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.