LINC00922: A Critical Oncogenic Long Non-Coding RNA Involved in Cancer Progression, Chemotherapy Resistance, and microRNA Regulation
Funding: This work was supported by the Qiantang Scholars Fund in Hangzhou City University (No. 210000-581835).
Jiahua Si, Chenhao Liang and Xueting Zhang contributed equally to this work and shared the co-first authorship.
ABSTRACT
LINC00922, a long non-coding RNA (lncRNA) located at chromosome 16q21, has emerged as a crucial regulatory molecule in cancer progression, chemotherapy resistance, and immune modulation. This lncRNA is predominantly localized in the cytoplasm, with limited expression in the nucleus and exosomes. Elevated LINC00922 expression levels are associated with poor patient prognosis across various cancer types. LINC00922 can regulate cancer cell behavior through complex gene regulatory networks, including interactions with several key signaling pathways, such as the Wnt, telomerase, and immune response pathways. Importantly, LINC00922 participates in multiple competing endogenous RNA axes, influencing the expression patterns of target microRNAs like miR-424-5p and miR-874-3p, which in turn regulate important cancer-related genes, such as TFAP2C and GDPD5 contributing to chemotherapy resistance. LINC00922 can also promote immune cell infiltration in tumors, with its high expression levels correlating with extensive immune cell presence, suggesting potential for cancer immunotherapy. Despite these promising findings, the upstream regulatory factors of LINC00922 remain poorly understood and further research is needed to fully uncover its clinical potential. In this review, we highlight the multifaceted roles of LINC00922 in cancer, emphasizing the need for future studies to explore its potential as a therapeutic target and diagnostic biomarker.
Abbreviations
-
- BC
-
- bladder cancer
-
- BrC
-
- breast cancer
-
- ceRNA
-
- competing endogenous RNA
-
- CRC
-
- colorectal cancer
-
- DFS
-
- disease-free survival
-
- DNMT
-
- DNA methyltransferase
-
- EMT
-
- epithelial-mesenchymal transition
-
- FZD
-
- frizzled
-
- GC
-
- gastric cancer
-
- HCC
-
- hepatocellular carcinoma
-
- LC
-
- lung cancer
-
- lncRNA
-
- long non-coding RNA
-
- miRNA
-
- microRNA
-
- OC
-
- ovarian cancer
-
- OS
-
- overall survival
-
- SaOS
-
- osteosarcoma
-
- SIRT3
-
- Sirtuin 3
-
- TCGA
-
- The Cancer Genome Atlas
-
- TNM
-
- tumor-node-metastasis
1 Introduction
Non-coding RNAs (ncRNAs) are a broad class of RNA molecules that play essential regulatory roles in cells despite not encoding proteins [1, 2]. Using their length, structure, and function, ncRNAs can be classified into various types, including long non-coding RNAs (lncRNAs), microRNAs (miRNAs) [3], circular RNAs (circRNAs) [4, 5], and Piwi-interacting RNAs (piRNAs) [6]. Each ncRNA type has distinct cellular functions and can regulate different aspects of gene expression. Among them, lncRNAs are defined as non-coding transcripts that are longer than 200 nucleotides and transcribed by RNA polymerase II [7]. With advances in epigenetics, the regulatory roles of lncRNAs in tumors have become increasingly clear, drawing significant research attention [8]. By competitively binding to miRNAs or directly interacting with protein-coding gene transcripts, lncRNAs can regulate the splicing, translation, and methylation of target genes, thereby modulating the expression levels and activity of their corresponding proteins [8]. One such lncRNA is LINC00922, which has a locus at 16q21. LINC00922 has attracted considerable interest because of its pivotal regulatory function. It is highly expressed in various cancer types and plays a critical role in tumor progression.
LINC00922 has eight alternative splicing variants, with LINC00922-203 being the representative transcript [9]. This lncRNA features a complex secondary structure, potentially serving as a binding site for target messenger RNA (mRNA) interaction or influencing cancer cell resistance mechanisms [10]. In various cancer cell lines, LINC00922 is predominantly localized in the cytoplasm, although a small portion is also found in the nucleus [11]. Extensive experimental evidence has confirmed that LINC00922 is abnormally expressed in numerous cancer types, with data from the GDC database further supporting this finding. LINC00922 operates within a complex gene regulatory network. The upstream factor TFAP2C binds to the promoter region of the gene encoding LINC00922, promoting its transcription. Additionally, LINC00922 can regulate the expression of downstream factors at both the pre-transcriptional and post-transcriptional levels, including NKD2, ETS1, and six competing endogenous RNA (ceRNA) axes. Interestingly, LINC00922 participates in a positive feedback loop, promoting its own accumulation in cancer cells via the LINC00922/miR-424-5p/TFAP2C axis in osteosarcoma (SaOS) cells [12].
In both in vitro and in vivo experiments, LINC00922 has been shown to promote several malignant processes, including tumor cell proliferation, the epithelial-mesenchymal transition (EMT), invasion, metastasis, and apoptosis. Furthermore, numerous independent studies have demonstrated that overexpression of LINC00922 is significantly associated with increased cancer cell resistance to chemotherapeutic drugs [12, 13]. Clinical data from patient follow-up evaluations confirm that high LINC00922 expression levels are correlated with poor cancer patient prognosis.
As an emerging focus of ncRNA research, LINC00922 has been found to play a pivotal role in cancer progression, holding significant potential for clinical applications in cancer diagnosis and treatment. The differential expression of LINC00922 across various cancers also positions it as a promising diagnostic biomarker. Additionally, its regulatory network influences multiple malignant processes, making LINC00922 a potential target for therapeutic intervention. In this review, we provide a comprehensive summary of the current research on LINC00922, highlighting its role in cancer biology, while also identifying existing gaps in knowledge. These insights will serve as a valuable resource for future studies and offer new perspectives for the clinical translation of LINC00922-based therapies.
2 LINC00922 Gene Characteristics
As illustrated in Figure 1a, LINC00922 is located at 16q21. A search of the Ensembl database (https://asia.ensembl.org/index.html, accessed August 17, 2024) revealed that LINC00922 has eight alternative splicing variants, with LINC00922-203 serving as the representative transcript [14]. Figure 1b shows the secondary structure prediction for LINC00922, as obtained from the lncHUB2 database (https://maayanlab.cloud/lncHUB2/, accessed August 17, 2024). The structure comprises multiple lncRNA-specific modules, including internal loops, stacked loops, multi-branched loops, hairpin loops, protruding loops, and external loops. Previous studies have demonstrated that such structures enable lncRNAs to regulate cancer cell behavior, including cell survival and drug resistance [15]. The complex LINC00922 secondary structure likely contributes to the regulation of malignant cancer behaviors.
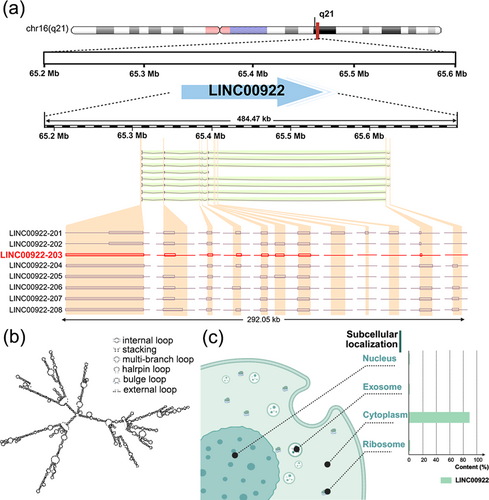
Characteristics and intracellular distribution of LINC00922. (a) Gene loci and transcripts of LINC00922, (b) Secondary structure of LINC00922, (c) Subcellular localization analysis of LINC00922.Source: Figure 1a and 1c were created by BioRender (biorender.com), Figure 1b was created by the lncHUB2 database (https://maayanlab.cloud/lncHUB2/).
LINC00922 is distributed in both the cytoplasm and nucleus. Subcellular fractionation and RT-PCR or fluorescent in situ hybridization (FISH) analysis of three different cancer cell lines have confirmed its cytoplasmic distribution in SaOS [12], ovarian cancer (OC) cell lines (ES-2 and SKOV3) [16], and gastric cancer (GC) cell lines (MKN-45 and HGC-27) [17]. However, FISH analysis of the breast cancer (BrC) cell line MCF-7 [18] and colorectal cancer (CRC) cell line HCT116 [19] revealed LINC00922 localization to be predominantly nuclear. To further clarify the distribution pattern, we consulted the lncLocator database, which confirmed that LINC00922 is predominantly cytoplasmic with some presence in the nucleus, as shown in Figure 1c [11].
Although LINC00922 has garnered attention as a newly discovered lncRNA, a deeper understanding of its genetic characteristics remains elusive. The nucleic acid properties of LINC00922 and its interactions with neighboring genes have yet to be fully characterized. Furthermore, the LINC00922 subcellular localization requires further experimental validation, with its nucleotide structure, secondary structure, and cellular distribution needing additional investigation to clarify its functional role in cancer progression.
3 Abnormal LINC00922 Expression in Multiple Cancer Types
As shown in Table 1, LINC00922 is significantly overexpressed in cancer samples compared with their corresponding normal tissues. LINC00922 is overexpressed in four cancer cell lines, including lung cancer (LC) [20], SaOS [12], OC [16, 25], and GC [17, 24]. Moreover, LINC00922 expression levels are significantly higher in highly invasive CRC cell lines compared with less invasive CRC cell lines [19], suggesting that LINC00922 contributes to the invasive ability of CRC cells. At the tissue level, LINC00922 is highly expressed in seven cancers: LC [20], SaOS [12], BrC [18, 26], bladder cancer (BC) [21], hepatocellular carcinoma (HCC) [22], OC [16, 25], and GC [17, 23, 24].
| Disease | Expression pattern | Sample type | Normal | Tumor | Reference |
|---|---|---|---|---|---|
| LC | Upregulated | Cell line | BEAS2B | A549, PC9, and H460 | [20] |
| Upregulated | Tissue | 52 matched normal tissues | 52 tumor tissues from LC patients | ||
| SaOS | Upregulated | Cell line | hFOB | Saos-2, U2OS, MG63, and MG63/DXR | [12] |
| Upregulated | Tissue | 40 normal adjacent tissue samples from SaOS patients | 40 tumor tissues from SaOS patients | ||
| BrC | Upregulated | Tissue | 113 matched normal tissues from TCGA database, 109 normal adjacent tissue samples from BrC patients, and six matched normal tissues from GSE26910 | 1104 tumor tissues from BrC patients from TCGA database, 109 tumor tissues from BrC patients, and six tumor tissues from BrC patients from GSE26910 | [18] |
| BC | Upregulated | Tissue | 20 normal adjacent tissue samples | 20 tumor tissues from BC patients | [21] |
| HCC | Upregulated | Tissue | 40 normal adjacent tissue samples from HCC patients | 40 tumor tissues from HCC patients | [22] |
| OC | Upregulated | Cell line | IOSE-80 | A2780, SKOV-3, and ES-2 | [16] |
| Upregulated | Tissue | 11 matched normal tissues from GSE74448 and matched normal tissues from TCGA database | 29 tumor tissues from OC patients from GSE74448 and tumor tissues from OC patients from TCGA database | ||
| GC | Upregulated | Cell line | GES-1 | MKN-45, HGC-27, AGS, and NCIN87 | [17] |
| Upregulated | Tissue | 40 normal adjacent tissue samples from GC patients | 40 tumor tissues from GC patients | ||
| Upregulated | Tissue | 210 normal adjacent tissue samples from GC patients from GTEx database and TCGA database, 32 normal adjacent tissue samples from GC patients from TCGA database, and 27 normal adjacent tissue samples from GC patients | 414 tumor tissues from GC patients from GTEx database and TCGA database, 375 tumor tissues from GC patients from TCGA database, and 27 tumor tissues from GC patients | [23] | |
| Upregulated | Cell line | GES-1 | HGC27, SGC7901, and AGS | [24] | |
| Upregulated | Tissue | 32 normal adjacent tissue samples from TCGA database | 375 tumor tissues from GC patients from TCGA database | ||
| CRC | Upregulated | Cell line | FHC | HCT116, LoVo, SW480, DLD-1, and SW620 | [19] |
- Abbreviations: BC, bladder cancer; BrC, breast cancer; CRC, colorectal cancer; GC, gastric cancer; GTEx, genotype-tissue expression; HCC, hepatocellular carcinoma; LC, lung cancer; OC, ovarian cancer; SaOS, osteosarcoma; TCGA, the cancer genome atlas.
| Disease | Location | Upstream factor | Target | Signaling pathway | Downstream factor | Reference. |
|---|---|---|---|---|---|---|
| LC | — | — | miR-204 | — | LINC00922/miR-204/HMGA2 | [20] |
| SaOS | Ccytoplasm | TFAP2C | miR-424-5p | — | LINC00922/miR-424-5p/TFAP2C | [12] |
| BrC | Nnucleus | — | NKD2 | Wnt | — | [18] |
| HCC | — | — | miR-424-5p | — | LINC00922/miR-424-5p/ARK5 | [22] |
| OC | Ccytoplasm | — | miR-361-3p | Wnt/β-catenin | LINC00922/miR-361-3p/CLDN1 | [16] |
| GC | — | — | miR-874-3p | — | LINC00922/miR-874-3p/GDPD5 | [13] |
| Ccytoplasm | — | miR-204-5p | — | LINC00922/miR-204-5p/HMGA2 | [17] | |
| CRC | Nnucleus | — | — | — | — | [19] |
- Abbreviations: BrC, breast cancer; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; LC, lung cancer; OC, ovarian cancer; SaOS, osteosarcoma.
| Disease | Effects in vitro | Cell line(s) | Effects in vivo | Cancer model | Reference |
|---|---|---|---|---|---|
| LC | EMT↑, invasion↑, and migration↑ | A549 and PC9 | — | — | [20] |
| SaOS | DXR-resistance↑ | MG63 | DXR-resistance↑ and tumor growth↑ | BALB/c nude mice | [12] |
| BrC | Viability↑, EMT↑, invasion↑, and migration↑ | MCF-7 | Tumor growth↑, lung metastasis↑, and liver metastasis↑ | BALB/c nude mice | [18] |
| BC | Invasion↑ and migration↑ | UMUC3 and RT4 | — | — | [21] |
| HCC | Viability↑, invasion↑, and migration↑ | SNU-182 and SK-Hep1 | — | — | [22] |
| OC | Proliferation↑, EMT↑, invasion↑, and migration↑ | ES-2 and SKOV3 | Tumor growth↑ | BALB/c nude mice | [16] |
| GC | Viability↑, invasion↑, migration↑, apoptosis↓, and DDP-resistance↑ | AGS and HGC-27 | Tumor growth↑ | BALB/c nude mice | [13] |
| Proliferation↑, invasion↑, migration↑, and apoptosis↓ | MKN-45 and HGC-27 | Tumor growth↑ and tumor infiltration↑ | Nude mice | [17] | |
| EMT↑, invasion↑, and migration↑ | MKN-45 | Tumor growth↑ and lung metastasis↑ | Nude mice | [23] | |
| Viability↑, proliferation↑, invasion↑, migration↑, apoptosis↓ | HGC27 | — | — | [24] | |
| CRC | Invasion↑ and migration↑ | HCT116 and LoVo | Lung metastasis↑ | BALB/c nude mice | [19] |
- Abbreviations: ↑: promote; ↓: inhibit; BC, bladder cancer; BrC, breast cancer; CRC, colorectal cancer; DDP, cisplatin; DXR, doxorubicin; EMT, epithelial-mesenchymal transition; GC, gastric cancer; HCC, hepatocellular carcinoma; LC, lung cancer; OC, ovarian cancer; SaOS, osteosarcoma.
| Disease | Sample size | LINC00922 expression pattern | Clinicopathological characteristic | Prognosis type | Reference |
|---|---|---|---|---|---|
| LC | 26 high expression LC patients versus 26 low expression LC patients | Upregulated | Later TNM stage, larger tumor size, and higher lymph node metastasis | Poor OS | [20] |
| SaOS | 40 SaOS patients | Upregulated | — | Poor OS | [12] |
| BrC | 54 high expression LC patients versus 54 low expression LC patients | Upregulated | Later TNM stage and larger tumor size | Poor OS | [18] |
| BC | 94 high expression BC patients versus 94 low expression BC patients, 93 BC patients from GSE31684 | Upregulated | — | Poor OS | [21] |
| OC | 178 high expression OC patients versus 503 low expression OC patients from TCGA database | Upregulated | — | Poor OS | [16] |
| GC | 184 high expression GC patients versus 173 low expression GC patients from TCGA database | Upregulated | — | Poor OS | [17] |
| 184 high expression GC patients versus 173 low expression GC patients from TCGA database | Upregulated | — | Poor OS, poor DFS, and poor PFS | [23] | |
| 188 high expression GC patients versus 187 low expression GC patients from TCGA database | Upregulated | Later T stage, later M stage, later histological grade, and different histological type | — | [23] | |
| 184 high expression GC patients versus 173 low expression GC patients from TCGA database | Upregulated | — | Poor OS and poor DFS | [24] |
- Abbreviations: BC, bladder cancer; BrC, breast cancer; DFS, disease-free survival; GC, gastric cancer; LC, lung cancer; OC, ovarian cancer; OS, overall survival; PFS, progression-free survival; SaOS, osteosarcoma; TCGA, the cancer genome atlas; TNM, tumor-node-metastasis.
These findings, confirmed by multiple independent studies, highlight that LINC00922 is overexpressed in various cancer types. To further explore the relationship between LINC00922 and cancer, we conducted bioinformatic analyses using The Cancer Genome Atlas (TCGA) and TARGET datasets [27] from the UCSC database (https://xenabrowser.net/, accessed August 7, 2024). This work revealed that LINC00922 is significantly overexpressed in seven tumor types, including BrC, acute lymphoblastic leukemia, pancreatic adenocarcinoma (PAAD), sarcoma (SARC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and ovarian serous cystadenocarcinoma (OV) (Supporting Information S1: Figure S1). In the breast invasive carcinoma dataset, the experimental results corroborated each other, showing significant overexpression of LINC00922. Conversely, LINC00922 was significantly underexpressed in four cancer types, including LUAD, LUSC, neuroblastoma, and testicular germ cell tumors (TGCT). These findings provide additional insights into the role of LINC00922 in cancer (for detailed information, see supplementary material).
4 Complex Regulatory Mechanism of LINC00922
As shown in Table 2, LINC00922 exhibits a complex regulatory mechanism and can modulate multiple genes at both the pre-transcriptional and post-transcriptional levels. At the pre-transcriptional level, LINC00922 can interact with DNA methyltransferases (DNMTs) and Sirtuin 3 (SIRT3), inhibiting the transcription of downstream proteins. At the post-transcriptional level, as illustrated in Figure 2a, LINC00922 suppresses the expression of several miRNAs, thereby promoting cancer progression. The binding sites of four miRNAs in LINC00922 are shown in Figure 2b. Additionally, Figure 3 demonstrates how LINC00922 can enhance its own expression levels in cancer cells through a positive feedback regulation mechanism [12].
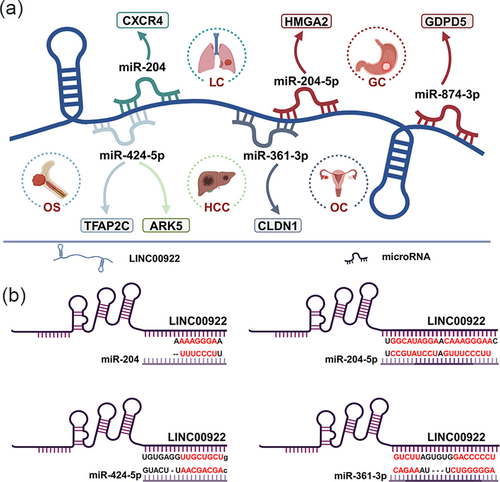
Post-transcriptional regulation of LINC00922. (a) LINC00922 participates in post-transcriptional regulation in five cancer types via six competing endogenous (ceRNA) axes, (b) Binding sites of LINC00922 with four microRNAs. GC, gastric cancer; HCC, hepatocellular carcinoma; LC, lung cancer; OC, ovarian cancer; SaOS, osteosarcoma.Source: All the figures were created by BioRender (biorender.com).
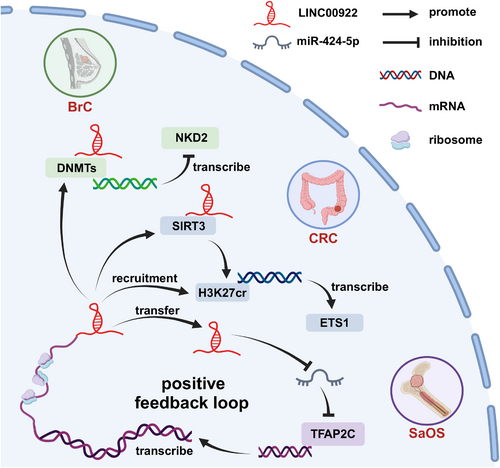
LINC00922 regulates multiple proteins. LINC00922 regulates the expression patterns of three proteins and enhances their levels in cancer cells through positive feedback regulation. BrC, breast cancer; CRC, colorectal cancer; SaOS, osteosarcoma.Source: All the figures were created by BioRender (biorender.com).
4.1 Upstream Factors Promoting LINC00922 Expression
DNA copy number amplification is a key contributor to the abnormal overexpression of lncRNAs in cancer cells [28]. TFAP2C, an upstream factor of LINC00922, is a transcription factor that can promote cancer cell proliferation, metastasis, and chemoresistance [29, 30]. According to the JASPAR database (https://xenabrowser.net/, accessed September 9, 2024), TFAP2C has two potential binding sites in the LINC00922 gene promoter region [12]. In the SaOS MG63 cell line, luciferase reporter assays confirmed that TFAP2C can bind to the LINC00922 promoter region and promote its expression [12]. Interestingly, LINC00922 can also enhance TFAP2C mRNA translation by inhibiting miR-424-5p expression [12]. This positive feedback loop results in the continuous accumulation of LINC00922 in cancer cells, with high LINC00922 expression patterns in MG63 cells, further suggesting that TFAP2C can promote its expression [12].
4.2 Pre-Transcriptional Regulation of LINC00922
Nucleotide modifications by specific enzymes play a crucial role in the pre-transcriptional regulation of lncRNAs [31]. For example, methylation and acetylation are important nucleotide modification mechanisms [32]. Mediating these modifications is a key way that lncRNAs can regulate gene expression at the pre-transcriptional level [33].
DNMTs, such as DNMT1, DNMT3A, and DNMT3B, are critical enzymes involved in transcriptional silencing and activation [34]. In BrC, LINC00922 can recruit DNMTs to the promoter region of NKD2, a tumor suppressor gene, by binding to DNMT1, DNMT3A, and DNMT3B [18]. This interaction leads to hypermethylation of the NKD2 promoter, inhibiting its expression and promoting cancer progression.
SIRT3, an NAD+-dependent acetylase involved in aging and aging-related diseases [35], plays a significant role in regulating cancer metastasis. High expression levels of H3K27cr, a styrylated histone protein, are correlated with CRC metastasis and poor patient prognosis [19]. H3K27cr and SIRT3 can both associate with the promoter region of ETS1, a transcription factor that promotes cancer metastasis [19]. H3K27cr and ETS1 cooperate to enhance the expression levels of cell adhesion molecules, facilitating cancer invasion and metastasis. In CRC, LINC00922 can bind to SIRT3, inhibiting its activity and recruiting H3K27cr to the ETS1 promoter. This action promotes the expression patterns of ETS1 and cell adhesion molecules, ultimately supporting cancer progression [19].
4.3 LINC00922 Promotes Six ceRNA Axes
LncRNAs can function as molecular sponges, also referred to as ceRNAs by exerting targeted regulatory effects on miRNAs [36]. A ceRNA axis describes the interaction in which a lncRNA inhibits a specific miRNA, thereby promoting the expression of the target genes of the miRNA [37].
Though this mechanism, LINC00922 can regulate multiple cancer-related pathways by participating in six ceRNA axes, including LINC00922/miR-204/CXCR4 in LC [20], LINC00922/miR-424-5p/TFAP2C in SaOS [12], LINC00922/miR-424-5p/ARK5 in HCC [22], LINC00922/miR-361-3p/CLDN1 in OC [16], and LINC00922/miR-874-3p/GDPD5 and LINC00922/miR-204-5p/HMGA2 in GC [13, 17]. Through these ceRNA axes, LINC00922 can promote cancer cell proliferation, invasion, metastasis, and survival.
5 LINC00922 in Wnt Signaling Pathway Regulation
The Wnt signaling pathway is a highly conserved and fundamental pathway involved in key cellular processes, such as proliferation, apoptosis, and stem cell differentiation [38]. This pathway is activated when Wnt proteins bind to specific surface receptors on the cell membrane [39]. The aggregation of Wnt and its receptors initiates the formation of a downstream destruction complex, which facilitates the nuclear translocation of β-catenin, driving the malignant progression of various cancers [39].
As illustrated in Figure 4, the classic Wnt pathway can be divided into two states: Wnt ON and Wnt OFF [40]. In the inactive state (Wnt OFF), a destruction complex composed of the scaffolding protein AXIN, tumor suppressor APC, and kinases CK1α and glycogen synthase kinase-3β(GSK-3β), binds to unphosphorylated β-catenin [40]. This complex facilitates β-catenin degradation rendering it inactive. In the activated state (Wnt ON), Wnt proteins bind to a protein heterodimer consisting of the transmembrane receptors frizzled (FZD) and LRP5/6, which triggers Wnt signaling pathway activation [40]. The activated FZD receptor binds to disheveled (DVL), leading to phosphorylation of LRP5/6. This results in the formation of the Wnt-FZD-LRP5/6 complex, which recruits AXIN1 and inhibits phosphorylation of β-catenin, promoting its nuclear translocation [40]. In the nucleus, β-catenin binds to transcription factors, driving the malignant progression of cancer cells [40]. In OC, LINC00922 inhibits miR-361-3p and activates the Wnt signaling pathway, which enhances β-catenin nuclear translocation and drives cancer cell proliferation [16]. NKD2, a tumor suppressor known to inhibit cell proliferation, invasion, and migration, has been shown to act as a negative regulator of the Wnt signaling pathway [18, 41]. In BrC, LINC00922 promotes Wnt signaling pathway activation by recruiting DNMT and reducing NKD2 gene expression [18]. Furthermore, Wnt signaling pathway activation facilitates the binding of nuclear β-catenin to transcription factors, which in turn drives c-Myc expression and promotes the EMT and tumor infiltration [18, 42].
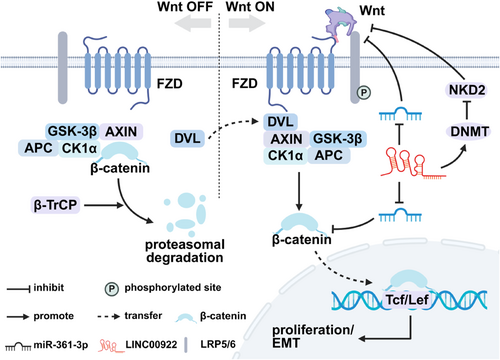
LINC00922 participates in the Wnt signaling pathway. LINC00922 promotes the malignant progression of breast and ovarian cancers by regulating the Wnt signaling pathway. CK1α, casein kinase 1 alpha; DNMT, DNA methyltransferase; DVL, disheveled; EMT, epithelial-mesenchymal transition; FZD, frizzled; GSK-3β, glycogen synthase kinase-3β; β-TrCP, beta-transducin repeat-containing proteins.Source: All the figures were created by BioRender (biorender.com).
6 LINC00922 Regulates Multiple Cellular Behaviors
As shown in Figure 5 and Table 3, both in vitro and in vivo experiments have confirmed that LINC00922 plays a key role in promoting malignant cancer progression. Through its involvement in various gene regulatory networks, LINC00922 can influence multiple cellular processes, including proliferation, the EMT, invasion, metastasis, and apoptosis. Additionally, tumor xenograft models have further validated the tumor-promoting effects of LINC00922.
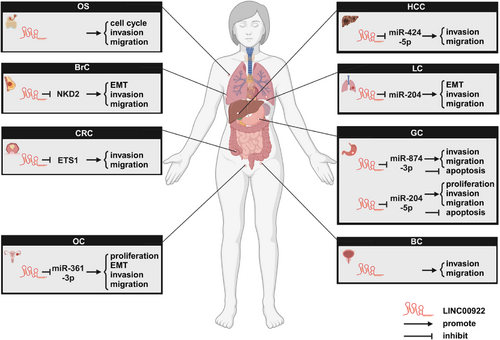
LINC00922 regulates cancer cell behavior. In eight cancer types, LINC00922 influences cellular behavior by participating in five competing endogenous (ceRNA) axes and inhibiting the expression patterns of two proteins. BC, bladder cancer; BrC, breast cancer; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; LC, lung cancer; OC, ovarian cancer; SaOS, osteosarcoma.Source: All the figures were created by BioRender (biorender.com).
6.1 LINC00922 Promotes Cancer Cell Proliferation and Tumor Growth in In Vitro Models
Cell proliferation is tightly regulated under normal physiological conditions. However, mutations in certain genes can dysregulate this process, ultimately leading to tumorigenesis [43, 44]. LINC00922 can enhance cell proliferation in OC and GC by modulating two ceRNA axes. In BrC, LINC00922 overexpression promotes hypermethylation of the NKD2 gene promoter, inhibiting NKD2 expression, which further accelerates MCF-7 cell proliferation [18]. In GC, LINC00922 overexpression enhances HGC-27 cell viability and promotes proliferation, though the specific pathway remains unconfirmed [24]. In another experiment, LINC00922 promoted the proliferation of GC cell lines (MKN-45 and HGC-27) via the LINC00922/miR-204-5p/HMGA2 axis [17]. In OC, LINC00922 can promote the proliferation of ES-2 and SKOV3 cells by engaging the LINC00922/miR-361-3p/CLDN1 axis [16].
In vivo experiments further supported the growth-promoting role of LINC00922 across multiple cancer types, including SaOS, BrC, OC, and GC. In BALB/c nude mice, LINC00922 could promote BrC tumor growth by inhibiting NKD2 expression. Moreover, in vivo experiments confirmed that LINC00922 could facilitate tumor growth in xenograft models of OC, GC, and other cancers through various ceRNA axes, including the LINC00922/miR-204/HMGA2 axis, LINC00922/miR-874-3p/GDPD5 axis, and LINC00922/miR-204-5p/HMGA2 axis [13, 17, 20]. However, the specific mechanisms involved in the SaOS and GC xenograft models remain to be elucidated.
6.2 LINC00922 Promotes the EMT Process of Cancer Cells
EMT describes the transformation of epithelial cells into mesenchymal-like cells, a process that significantly contributes to cancer progression, metastasis, and therapeutic resistance [45, 46]. In OC and GC, cells overexpressing LINC00922 exhibited elevated E-cadherin and reduced vimentin expression levels, which are indicative of EMT activation [16, 23].
LINC00922 can facilitate the EMT in LC and OC by inhibiting key regulatory proteins and participating in ceRNA axes. In BrC, LINC00922 promotes the EMT by downregulating NKD2 expression levels [18]. Expression of CXCR4, a G protein-coupled receptor implicated in cancer metastasis [47], is also upregulated by LINC00922 in LC, where it can promote the EMT via the LINC00922/miR-204/CXCR4 axis [20].
6.3 LINC00922 Enhances Cancer Cell Invasion and Metastasis
Cancer cell invasion and metastasis occur when cells break through the basement membrane, enter the bloodstream or lymphatic system, and spread throughout the body [48]. LINC00922 can promote invasion and metastasis in six cancer types (BC, CRC, LC, HCC, OC, and GC) by reducing NKD2 expression or participating in four ceRNA axes. In three independent experiments in BC and GC, overexpression of LINC00922 was shown to enhance cancer cell invasion and metastasis.
In CRC, LINC00922 binds to SIRT3, a protein that interacts with H3K27cr, leading to increased H3K27cr levels in the ETS1 promoter region [19]. This modification enhances ETS1 expression patterns, driving the invasion and migration of CRC cells (HCT116 and LoVo) [19].
Moreover, LINC00922 can promote cancer invasion and migration via ceRNA axes, including the LINC00922/miR-204/CXCR4 axis in LC, LINC00922/miR-424-5p/ARK5 axis in HCC, and LINC00922/miR-204-5p/HMGA2 axis in GC [13, 17].
In vivo experiments have also confirmed the tumor-promoting effects of LINC00922 on invasion and metastasis using BrC, GC, and CRC xenograft models. LINC00922 overexpression in GC and CRC xenografts could promote lung metastasis, while it promoted both lung and liver metastasis by inhibiting NKD2 transcription in BrC [18, 19, 23].
6.4 LINC00922 Inhibits Apoptosis in Cancer Cell Lines
Apoptosis is a key cellular process that eliminates damaged or abnormal cells [49]. In cancer, resistance to apoptosis significantly contributes to tumor progression [50]. In two independent GC experiments, overexpression of LINC00922 inhibited apoptosis in cancer cell lines [13, 17].
In these studies, LINC00922 promoted cancer progression by participating in two distinct ceRNA axes. For example, in the LINC00922/miR-874-3p/GDPD5 axis, LINC00922 could inhibit apoptosis in the AGS and HGC-27 GC cell lines [13]. Additionally, through the LINC00922/miR-204-5p/HMGA2 axis, LINC00922 can also inhibit apoptosis in the MKN-45 and HGC-27 cell lines [17].
7 LINC00922 Increases Cancer Cell Resistance to Chemotherapy Drugs
Chemotherapy is a cornerstone of cancer treatment, designed to inhibit tumor cell proliferation and systemic metastasis. However, the development of chemotherapy resistance in cancer cells significantly reduces the therapeutic efficacy of these drugs [51]. As shown in Figure 6, LINC00922 has been found to increase chemotherapy resistance in cell lines of two cancer types, SaOS [12] and GC [13], by participating in ceRNA networks.
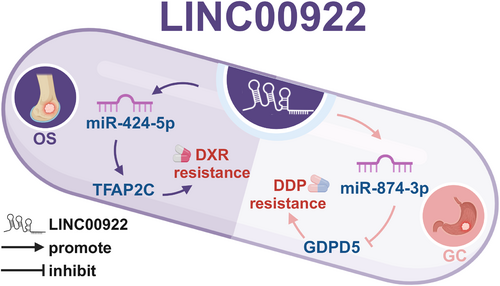
LINC00922 enhances chemotherapy resistance in cancer cells. LINC00922 participates in two competing endogenous (ceRNA) axes, which can enhance chemotherapy resistance in two cancer types (SaOS and GC). DDP, cisplatin. DXR, doxorubicin; GC, gastric cancer; SaOS, osteosarcoma.Source: All the figures were created by BioRender (biorender.com).
TFAP2C, a member of the transcription factor AP-2 family, is associated with drug resistance in several cancers [30]. High TFAP2C expression levels are correlated with increased drug resistance in CRC cells [30]. GDPD5, a choline-producing enzyme, has been linked to resistance to 5-FU in CRC [52]. In SaOS, LINC00922 overexpression increases TFAP2C expression levels in the MG63 cell line via the LINC00922/miR-424-5p/TFAP2C axis, thus enhancing resistance to the chemotherapy drug doxorubicin (DXR) [12]. In vivo studies using BALB/c nude mice further confirmed that LINC00922 can boost DXR resistance in cancer cells through this ceRNA axis [12]. Similarly, in GC, LINC00922 overexpression elevates GDPD5 expression levels in the AGS and HGC-27 cell lines through the LINC00922/miR-874-3p/GDPD5 axis, thereby increasing resistance to DDP [13].
8 LINC00922 Increases Immune Cell Infiltration in Cancer Patients
Immune cell-based therapies are emerging as a promising approach for cancer treatment, providing new avenues for tumor management [53]. The presence and activity of various immune cell types in the tumor microenvironment are closely associated with cancer progression and metastasis [19]. Using the ssGSEA algorithm in the GSVA package, we observed that LINC00922 expression levels positively correlate with the infiltration of multiple immune cell types, including cytotoxic T cells, dendritic cells (DC), immature DCs, eosinophils, macrophages, mast cells, neutrophils, natural killer (NK) cells, plasmacytoid DCs, T follicular helper cells, γδ T cells, Th1 cells, and regulatory T cells (Treg) [23]. However, high LINC00922 expression levels were negatively correlated with Th17 cell infiltration [23]. Further analysis revealed significant differences in immune cell infiltration patterns between high- and low-expression LINC00922 GC samples, reinforcing the association between LINC00922 expression levels and immune cell content in these patients.
To further confirm the relationship between LINC00922 and immune cells, we analyzed the TCGA Pan-Cancer dataset extracting LINC00922 expression data across samples from 30 different cancer types. Using the EPIC method to assess immune cell infiltration, we found a positive correlation between LINC00922 expression levels and immune cell infiltration in multiple cancer types (Supporting Information S1: Figure S2).
9 High LINC00922 Expression Levels Predict Poor Prognosis
As shown in Table 4, increased LINC00922 expression levels are strongly associated with poor prognosis in various cancer types. Prognostic analyses have revealed that patients with elevated LINC00922 expression levels tend to exhibit advanced pathological features, such as later tumor-node-metastasis (TNM) stages, higher T stages, poorer histological grades, larger tumors, and more frequent lymph node metastasis. High LINC00922 expression levels are also linked to worse overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS) in numerous cancers, such as LC [20], SaOS [12], BrC [18], OC [16], GC [17, 23, 24], CRC [19], and BC [21].
In LC, LINC00922 expression patterns can predict poor OS, with higher levels correlating with later TNM stages, larger tumor size, and increased lymph node metastasis [20]. In SaOS, LINC00922 expression levels are linked to poor OS [12]. In BrC, high LINC00922 expression patterns correlate with poor OS, advanced TNM stages, and larger tumor size [18]. In OC, LINC00922 expression levels are predictive of poor OS [16]. In GC, LINC00922 expression can predict poor OS, DFS, and PFS, with higher expression levels associated with more advanced T and M stages, worse histological grades, and different histological types [23]. Additionally, other studies have shown that LINC00922 expression levels correlate with poor OS and DFS in GC [24] and are a predictor of poor OS in CRC [19].
Moreover, H3K27cr, a downstream factor of LINC00922, is highly expressed in tumors with more advanced stages and metastatic features, further confirming the prognostic significance of LINC00922 expression in predicting cancer progression [19].
10 Discussion
NcRNAs encompass a diverse range of molecules, including lncRNAs, miRNAs [3], circRNAs [4, 5], and piRNAs [6]. Previous studies have identified hundreds of ncRNAs involved in RNA maturation, transcriptional regulation, chromatin remodeling, and post-transcriptional modifications. Each ncRNA subtype plays a distinct regulatory role in gene expression and contributes to various aspects of cancer development. Over the past decade, lncRNA research has rapidly advanced, uncovering their remarkable properties. LncRNAs are now recognized as crucial players in numerous cellular processes, including the cell cycle, differentiation, and metabolism [8]. These molecules exert their functions through various mechanisms, such as regulating transcription, epigenetic modifications, controlling protein and RNA stability, and influencing the translation and post-translational modifications of RNA or proteins [54]. Consequently, targeting lncRNAs has emerged as a promising strategy for cancer therapy [55].
The gene encoding LINC00922, a recently discovered lncRNA, is located on chromosome 16 at 16q21. LINC00922 is predominantly localized in the cytoplasm, with smaller amounts found in the nucleus, ribosomes, and exosomes. Clinical data have shown that high LINC00922 expression levels are closely associated with various pathological features of malignant tumors and poor patient prognosis. Studies comparing normal and cancerous tissues at both the cellular and tissue levels have confirmed that LINC00922 expression levels are significantly elevated in many cancers, indicating that this molecule is strongly correlated with cancer development. Our pan-cancer analysis using data from the GDC database further supports this observation, revealing significant LINC00922 expression differences across diverse cancer types. However, current studies have mainly performed single-cell and tissue analyses, with limited research on LINC00922 expression patterns in common clinical body fluids. Currently, there is a lack of clinical data and experimental validation to confirm the feasibility and accuracy of LINC00922 as a potential therapeutic target and nuclear diagnostic biomarker in clinical applications. Future studies should aim to collect more comprehensive clinical datasets and integrate larger, diverse databases to analyze LINC00922 expression differences between cancer patients and healthy individuals across various tissues, cells, blood, and body fluids. This approach will help further validate the reliability of using LINC00922 as a clinical biomarker for cancer diagnosis. Thus, although LINC00922 holds promise as a clinical target and biomarker, further experiments are needed to fully assess its potential.
LINC00922 can regulate cancer cell functions by participating in complex gene regulatory networks. LINC00922 is known to be regulated by the activating protein TFAP2C in cancer cells [12]. Interestingly, it forms a positive feedback loop via the LINC00922/miR-424-5p/TFAP2C axis, continuously elevating LINC00922 expression levels in cancer cells, thereby promoting malignant progression [12]. Additionally, LINC00922 can directly target NKD2 [18] and five miRNAs, inhibiting their expression patterns. NKD2 is a negative regulator of Wnt signaling that is crucial for controlling activation of this pathway [18, 56]. By suppressing NKD2, LINC00922 can promote Wnt pathway activation. LINC00922 can also indirectly enhance the expression of the telomerase regulator ETS1 by recruiting H3K27cr in the nucleus, thereby facilitating cancer cell invasion and metastasis [19]. However, the downstream regulatory mechanisms of LINC00922 are still incompletely understood, as its upstream regulatory factors remain largely unexplored. LINC00922 has been shown to suppress the expression of multiple miRNAs, thereby modulating cancer cell proliferation, invasion, and metastasis through several ceRNA axes. Additionally, experimental evidence has indicated that LINC00922 can activate the Wnt signaling pathway by inhibiting its downstream targets, NKD2 and miR-361-3p, though the exact regulatory mechanism remains unclear. Future studies using CRISPR technology or western blot analysis may help clarify this pathway. While TFAP2C has been identified as an upstream factor, other proteins involved in regulating LINC00922 remain unidentified. LncRNAs exhibit complex regulatory mechanisms with diverse and intricate upstream factors [57]. Future studies should focus on elucidating these regulatory mechanisms through more detailed experiments. For future exploration of upstream regulators of LINC00922, ChIP-seq technology could be employed to identify transcription factors, along with methylation analysis and promoter region variation studies. Additionally, epigenetic modifications, such as DNA methylation and histone modifications, may be involved in regulating LINC00922 expression.
Moreover, LINC00922 is involved in key cellular processes, such as proliferation, the EMT, invasion, metastasis, and apoptosis regulation. Both in vivo and in vitro experiments have shown that LINC00922 can regulate cancer cell behavior by participating in five ceRNA axes and other mechanisms, significantly impacting the development of various cancers. In GC, multiple independent studies have confirmed that LINC00922 promotes malignant behavior in GC cell lines through different pathways. Although these findings have greatly expanded our understanding of the LINC00922 regulatory roles, the precise mechanisms by which this lncRNA contributes to cancer progression remain unclear. Although LINC00922 has been shown to promote cancer cell malignant behavior, the detailed regulatory pathways are still unknown.
Prior work has suggested that LINC00922 can promote cancer progression in multiple malignancies, such as GC, by participating in ceRNA axes [17]. However, its role in certain tumor types, including how it can promote lung metastasis in CRC, remains unclear [19]. Future research should investigate cancer-related tumor suppressors or oncogenic factors as entry points to better understand the full regulatory scope of LINC00922 using proteomics to explore its relationship with cancer-related cytokines. Numerous in vitro studies and some in vivo experiments have demonstrated the involvement of LINC00922 in various malignancies, with bioinformatics analyses using public datasets suggesting that LINC00922 is potentially related to cellular immunity across multiple cancer types. However, direct evidence supporting its role in inhibiting cancer progression remains insufficient. The precise function of LINC00922 in regulating tumor development and the underlying mechanisms through which it influences cancer progression warrant further investigation. Future research should prioritize the collection of broader and more robust clinical data and conduct in-depth integrative analyses using multiple databases. This will help elucidate the critical role of LINC00922 in cancer biology and assess its viability as a therapeutic target in clinical settings.
In cancer treatment, elevated LINC00922 expression levels are associated with increased cancer cell resistance to chemotherapy drugs [12, 13]. Data analysis has suggested that high LINC00922 expression levels correlate with extensive immune cell infiltration in GC tissues. To further explore the connection between LINC00922 and immunity, we analyzed data from the TCGA database, confirming that LINC00922 expression patterns are positively associated with immune cell infiltration in multiple cancer types. This suggests that LINC00922 may serve as a new target for both cancer chemotherapy and cellular immunotherapy. However, the precise relationship between LINC00922 and immune cell infiltration in cancer requires further experimental validation. Immune resistance and targeted drug resistance are common mechanisms of tumor immune evasion in clinical cancer treatment [58, 59]. However, the potential roles of LINC00922 in these processes have not yet been experimentally verified. Future studies should explore how this lncRNA is specifically involved in drug resistance mechanisms. Additionally, the impact of LINC00922 on immune cell-targeted therapies has yet to be verified, representing a potential avenue for future research. Targeted therapy and immunotherapy are key approaches in cancer treatment. A deeper understanding of the relationship between LINC00922 and targeted therapies will provide critical insights into its potential clinical applications [58, 59]. However, its roles in regulating immunotherapy resistance and targeted drug resistance have not yet been experimentally validated. Future studies should investigate the specific mechanisms underlying LINC00922-mediated drug resistance. Furthermore, clinical data and patient follow-up studies have shown that high LINC00922 expression levels can predict poor prognosis in several cancer types, suggesting that this lncRNA could serve as a novel prognostic biomarker for cancer patients. Although LINC00922 has been associated with poor prognosis in a limited number of cancers, its prognostic significance in other malignancies remains uncertain. While existing studies have confirmed its high expression patterns in multiple cancer types, there are insufficient data to establish a direct correlation between high LINC00922 expression levels and poor prognosis in cancer patients. Future research should incorporate extensive clinical data and correlation analyses to validate its prognostic potential.
In conclusion, this article summarizes the aberrant LINC00922 expression patterns in various cancers as well as explores its associated gene regulatory network and effects on various tumor cell biological behaviors. We also examined the impacts of LINC00922 overexpression on chemotherapy drug responses and its prognostic value. As a potential oncogenic lncRNA, LINC00922 is poised to become a new diagnostic biomarker and therapeutic target, warranting further in-depth investigation.
Author Contributions
Jiahua Si: conceptualization (equal), data curation (equal), formal analysis (equal), visualization (equal), writing – original draft (equal). Chenhao Liang: data curation (equal), formal analysis (equal), visualization (equal), writing – original draft (equal). Xueting Zhang: data curation (equal), visualization (equal), writing – original draft (equal). Hening Xu: visualization (equal), writing – original draft (equal). Xinming Su: writing – original draft (equal). Luqing Liu: writing – original draft (equal). Ruyi Yuan: writing – original draft (equal). Yuemin Ding: conceptualization (equal), supervision (equal), writing – review and editing (equal). Shiwei Duan: conceptualization (equal), funding acquisition (equal), project administration (equal), writing – review and editing (equal).
Acknowledgments
The authors have nothing to report.
Ethics Statement
The authors have nothing to report.
Consent
The authors have nothing to report.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data supporting the findings of this study are openly available from multiple sources: the Ensembl database (reference number 10.1186/s13059-016-0974-4), the lncHUB2 database (reference number 10.1093/database/baad009), the NPInter database (reference number 10.1093/nar/gkz969), and the UCSC database (reference number 10.1093/nar/gkaa1070). Additional supporting data are provided in the Supporting Information S1 of this article.




