Multifaceted role of TRIM28 in health and disease
Abstract
The TRIM (tripartite motif) family, with TRIM28 as a key member, plays a vital role in regulating health and disease. TRIM28 contains various functional domains essential for transcriptional regulation, primarily through its interaction with KRAB-ZNF proteins, which influence chromatin remodeling and gene expression. Despite extensive research, the precise mechanisms by which TRIM28 impacts health and disease remain elusive. This review delves into TRIM28’s multifaceted roles in maintaining health, contributing to a variety of diseases, and influencing cancer progression. In cancers, TRIM28 exhibits a dual nature, functioning as both a tumor promoter and suppressor depending on the cellular context and cancer type. The review also explores its critical involvement in processes such as DNA repair, cell cycle regulation, epithelial-to-mesenchymal transition, and the maintenance of stem cell properties. By uncovering TRIM28’s complex roles across different cancers and other diseases, this review underscores its potential as a therapeutic target. The significance of TRIM28 as a versatile regulator opens the door to innovative therapeutic strategies, particularly in cancer treatment and the management of other diseases. Ongoing research into TRIM28 may yield key insights into disease progression and novel treatment options.
1 INTRODUCTION
The proteins encoded by the tripartite motif (TRIM) gene family are diverse and involved in a range of biological processes. These proteins comprises a coiled-coil region, one or two B-box motifs, and an extremely intriguing new gene (RING) domain—collectively called the RING finger, B-box, and coiled-coil (RBCC) motif 1—is present in these proteins.1
The TRIM superfamily of proteins is distinguished by the remarkably conserved sequence of domains within the RBCC motif. This conservation across different species highlights the RBCC motif as the key defining feature of the superfamily. Even in TRIM family members that lack one of the RBCC domains, the remaining domains maintain their order and spacing, further emphasizing the motif's critical role.2 Due to the pivotal role of the TRIM family in posttranslational protein modification, cellular signaling pathways, and gene regulation, it is unsurprising that different members of the TRIM family exhibit a range of effects on cell behavior. These impacts encompass the control of cell multiplication, movement, and infiltration.3, 4
As noted, the TRIM family is extensive, with each member contributing uniquely to both disease and health. Mutations in specific TRIM genes, such as MID1, TRIM32, and TRIM37, are linked to hereditary genetic disorders, including X-linked Opitz G/BBB syndrome, limb-girdle muscular dystrophy type 2H, and Mulibrey nanism, respectively. Many TRIM proteins respond to interferon (IFN), with some serving as downstream effectors in innate immune responses to retroviruses and other viral infections. For example, TRIM proteins exhibiting anti-HIV activity can disrupt various stages of the virus's life cycle, including preintegration (e.g., TRIM5), transcription (e.g., TRIM22, TRIM32), and assembly (e.g., TRIM22, TRIM15). Additionally, other TRIM genes—such as TRIM21, TRIM25, TRIM27, TRIM30, and TRIM32—function downstream of IFN and pathogen-recognition receptors, playing a role in modulating innate immune responses to bacterial and viral infections through the activation of IRF3, IRF7, and NF-κB. Furthermore, TRIM21 and TRIM68 are significant targets of autoantibodies in individuals with autoimmune diseases, including systemic lupus erythematosus and Sjögren's syndrome.5
We focused our investigation on TRIM28 due to its significant involvement in various diseases. Addressing this issue is crucial because cancer is one of the main causes of death. In this paper, we not only conducted a bioinformatics analysis and examined the relationship between TRIM28 and cellular processes in health and disease, particularly in cancer, but also provided a valuable update on this subject.
2 NORMAL DEVELOPMENT/HEALTH OF TRIM28
TRIM28 is an important protein for normal development and health. It is essential in embryonic development, cell differentiation, genomic stability, and the maintenance of adult tissue homeostasis.6 TRIM28 regulates genes associated with pluripotency, ensuring that embryonic stem cells (ESCs) maintain their pluripotent state until differentiation is properly signaled. It also maintains epigenetic marks, such as DNA methylation, which are essential for stable gene silencing during differentiation. Additionally, TRIM28 contributes to genomic stability by silencing transposable elements (TEs) and repairing double-strand breaks. Moreover, it is extremely important in brain development, notably in the regulation of neural progenitor cells. TRIM28 also regulates immune system genes and maintains cellular homeostasis in adult tissues by controlling gene expression and protecting against genomic instability. Furthermore, it plays a role in metabolic processes, such as adipogenesis, which controls the balance between energy storage and expenditure.7
2.1 The structure and function of TRIM family
A RING finger domain, one or more B-box domains, and a coiled-coil region are the three primary motifs that make up the TRIM family. These motifs, typically located near the protein's N-terminal, form the tripartite motif that gives the family its name. The RING finger domain, a type of zinc finger motif, binds zinc ions through conserved histidine and cysteine residues. This domain is crucial for the E3 ubiquitin ligase activity of many TRIM proteins, enabling them to transfer ubiquitin molecules to substrate proteins, marking them for degradation by the proteasome. Zinc-binding motifs are also known as B-box domains, and TRIM proteins typically contain one or two of these domains (B-box 1 and B-box 2). They play roles in protein–protein interactions (PPIs) and stabilization of the TRIM protein structure. The formation of higher-order oligomers or homo- or hetero-dimers is facilitated by the coiled-coil region. Since dimerization or oligomerization frequently influences the biological activity of TRIM proteins, this region is essential to their function. TRIM proteins’ C-terminal region exhibits greater variability, containing different domains depending on the specific protein. These domains include PRY/SPRY, COS, FIL, MATH, and B30.2, which are involved in PPIs, fibronectin type III-like functions, associations with intermediate filaments, PPIs, and recognition of specific protein substrates, respectively. Furthermore, this protein family plays a role in a number of biological processes, such as cancer, immunological response, cell division and proliferation, ubiquitination, and protein degradation.8, 9
2.2 The structure and function of TRIM28
TRIM28, along with three other TRIM proteins—TRIM24, TRIM33, and TRIM66—constitutes the TIF1 family.10, 11 The domains located in the N-terminus of TRIM28 are known as the RBCC domain, also referred to as the TRIM domain.9 The RING finger domain is distinguished by its cysteine-rich sequence, being essential to its interaction with the Kruppel-associated box (KRAB) domain, which is present in many KRAB-zinc finger (KRAB-ZFP).12, 13
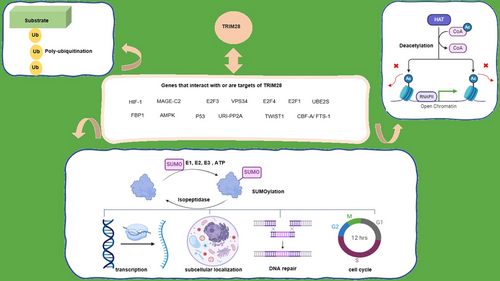
The core segment of the TRIM28 protein harbors the PxVxL pentapeptide domain, facilitating its binding with HP1.14 Furthermore, akin to other TIF1 proteins, TRIM28 also possesses the characteristic TSS at its central region, comprising a 25-amino acid motif abundant in tryptophan and phenylalanine residues. At the carboxyl (C) terminus of TRIM28, the plant homeodomain (PHD) finger and bromodomain reside, serving to recruit components of NuRD, including the histone deacetylase complex and the H3K9-specific methyltransferase SETDB1, thereby promoting chromatin condensation. Both the PHD finger and the RING domain, located here, exhibit a structure rich in cysteine and histidine residues, featuring a consensus Cys4–His–Cys3 motif spanning 50–80 residues. The collective presence of the PHD, bromodomain, and PxVxL domain is proposed to augment the formation of condensed heterochromatin, marked by increased affinity for HP1 binding.14 TRIM28 is subject to regulation at multiple levels, encompassing gene transcription, posttranscriptional translation, and posttranslational modifications. For instance, WDR4 is capable of inducing the transcription of the TRIM28 gene.15 ZBRK1/ZNF350 inhibits TRIM28 transcription by repressing the activity of its promoter.16 TRIM28’s suppressive function involves recruiting SETDB1 and the NuRD complex, as well as interacting with the HP1 protein. Notably, TRIM28 also acts as an intramolecular E3 SUMO ligase.17 TRIM28 possesses ubiquitin E3 ligase activity, facilitating substrate degradation, and enhances the SUMOylation of substrates. SUMOylation is another key post-translational modification that involves the addition of a small ubiquitin-like modifier.18 Like ubiquitination, SUMOylation involves a cascade of enzymatic steps, including E1 activation, E2 conjugation, and E3 ligation, to attach SUMO molecules to target substrates.18 Although SUMO E3 ligases are not essential for the SUMOylation process, their dysregulation can significantly impact substrate SUMOylation, contributing to both normal and cancer development.19 For normal development, proper SUMOylation is essential. In order to control gene expression, signaling pathways, and developmental processes, SUMO E3 ligases are essential. Because improper SUMOylation can interfere with cellular differentiation, proliferation, and tissue formation, dysregulation of SUMOylation resulting from aberrant E3 ligase function can cause developmental defects.20 The transcriptional repression activity of TRIM28 is dependent on the SUMOylation of three specific lysine residues (K554, K779, and K804). Importantly, within the TRIM28 molecule, the PHD and bromodomain regions act as SUMO E3 ligases, with the PHD domain binding to UBC9 (the SUMO E2 enzyme) and working together to enhance the SUMOylation of TRIM28.21
2.3 TRIM28 and transcriptional coregulation
Being a transcriptional corepressor, the TRIM28 protein is essential for KRAB-ZNF proteins to perform their repressive function.22 In brief, KRAB-ZNF proteins, anchored by their zinc finger domains to specific DNA recognition motifs, recruit the TRIM28 protein. TRIM28 then serves as a scaffold for various heterochromatin-inducing factors.23 Subsequently, the PHD-mediated SUMOylation of the bromodomain occurs, which then assists in recruiting SETDB1 and NuRD complex proteins. This sequential process ultimately leads to histone deacetylation.21 Furthermore, the HP1 protein interacts with TRIM28 at the PxVxL motif and associates with the H3K9me3 mark, thereby enhancing the stability of the TRIM28-containing complex that is bound to KRAB-ZNF.24 Changes in chromatin organization, prompted by TRIM28 recruitment to particular genomic loci through synthetic constructs, result in the suppression of transcription from RNA polymerase I, II, and III promoters.25 TRIM28 plays a role in stabilizing the pausing of RNA polymerase II near the transcriptional start site in various inactive genes.26 The modulation of Pol II pausing is contingent upon the phosphorylation status of TRIM28. When TRIM28 is phosphorylated at Ser824, it facilitates the release of Pol II from pausing, leading to the rapid transcription of target genes. Furthermore, studies have shown that TRIM28 regulates the transcription of a subset of lncRNAs in mammalian cells through a similar mechanism involving Pol II pausing at their transcriptional start sites.26 Given the abundance of noncoding RNAs encoded by mammalian genomes, which surpass mRNA genes by a significant margin, it is reasonable to speculate on the extensive role of TRIM28 in the regulation of genome-wide transcription.27 Figure S1 represents the PPI between TRIM28 and other proteins. The strong interaction between TRIM28 and the ZNF and CBX families is seen in Figure S1. The data indicate a robust association between members of the ZNF family and TRIM28, an essential transcriptional regulator implicated in DNA binding and regulation of gene expression. This interaction is particularly relevant in the context of both normal development and disease, particularly in cancer since dysregulation of ZNF proteins has been related to carcinogenesis through their effect on tumor suppressor genes and oncogenes. TRIM28 is essential for the creation and preservation of epigenetic marks when it forms a complex with ZNF proteins. It plays a role in the processes that preserve genomic stability by forming heterochromatin and silencing TEs. Normal cellular differentiation and the expression of genes specific to particular tissues during development depend on this epigenetic regulation. Diseases and disorders related to development can result from disruptions in these processes.7
Significant ties to the Chromobox (CBX) family, which are crucial components of the polycomb repressive complexes that maintain the repression of gene transcription, are also highlighted in the image. Because they maintain tumor suppressor genes in their repressed form, members of the CBX family are essential to the initiation and progression of cancer. These interactions demonstrate how essential TRIM28 is for regulating a network of protein partners that improve gene silencing and chromatin remodeling. This draws attention to the role of the protein in biological processes like differentiation and development, as well as any possible links to cancer. TRIM28 may affect the epigenetic landscape of cancer cells by controlling the relationships between various protein families, so fostering an environment that is favorable to malignancy and resistance to treatment.
2.4 TRIM28 and DNA damage repair
One of the many cytotoxic DNA lesions that can trigger the cellular response to DNA damage is double-strand breaks. Once DNA damage is detected, TRIM28 aids in chromatin remodeling near the break location. This is accomplished by TRIM28 through the use of histone-modifying enzymes, including HDACs.28 Additionally, the activation of ataxia telangiectasia mutated (ATM) kinase, belonging to PIKK family, is essential for addressing this form of damage.29 The ATM kinase phosphorylates the TRIM28 protein at Ser824 located within the C-terminus and at Ser473 in close proximity to the HP1 binding domain.30, 31 The process of TRIM28 phosphorylation marks one of the initial stages in the cellular response to DNA damage.31 Elimination of the Ser824 phosphorylation site on TRIM28 results in the inability to induce chromatin decondensation in response to double-strand breaks within DNA. Additionally, it increases the cells’ susceptibility to agents that induce double-strand breaks.30 Reducing TRIM28 levels or introducing a constitutive Ser824 phosphorylation mutation (TRIM28–S824D mutant) results in persistent chromatin relaxation. These results underscore the importance of chromatin relaxation as a core mechanism in the DNA damage response and highlight its key mediators.30 As evidenced in melanoma cancer cells, the MAGE-C2 protein has the capacity to trigger ATM-dependent phosphorylation of TRIM28 at Ser824, thereby favoring DNA damage repair mechanisms over apoptosis.32 MAGEC2 enhances the coprecipitation of TRIM28 with ATM, which is essential for the increased phosphorylation of TRIM28. This heightened phosphorylation level, in turn, enhances DNA damage repair, thereby promoting tumor progression.32 TRIM28 operates in inhibiting E2F transcription factor 1 (E2F1) and may act as a partial backup mechanism to impede E2F1-induced apoptosis in cancer cells.33
TRIM28 has been linked to the regulation of genes involved in neuronal development and function. Accumulation of DNA damage in neurons, a hallmark of neurodegenerative diseases such as Huntington's and Alzheimer's, can be facilitated by abnormalities in TRIM28 activity.34 TRIM28 interacts with RE-1-silencing transcription factor and huntingtin protein mutated, resulting in changes in transcriptional repression and chromatin remodeling, which could be an effective factor in increasing DNA damage and neurodegeneration.35
3 THE ROLE OF TRIM28 IN DISEASES
Although the significance of epigenetics was initially recognized for its role in tissue development, growing evidence now indicates that it is equally crucial in the onset and progression of numerous common diseases. Numerous novel risk factors have been discovered through population-based epigenetic epidemiological studies examining the impact of epigenetic changes on common diseases; however, this relatively nascent field continues to confront several distinct challenges. Among these, TRIM28 has been identified as a pivotal gene due to its regulatory influence on epigenetic processes. Variations in the expression or function of TRIM28 can aid in the development of a wide variety of illnesses, underscoring its vital importance in both health and disease contexts.
3.1 The TRIM family in cancer pathogenesis
Numerous genes within the TRIM family exhibit notable modifications across different cancer types.3, 4 TRIM family proteins associated with some various cancers are reported in Table 1. In specific cases, the expression levels of TRIMs can function as biomarkers and prognostic indicators for cancer.36 KRAB-associated protein 1 (a large multidomain protein (110 kDa), KAP1, also referred to as TRIM28 and TIF1b), initially garnered significant interest in 1996.37 Subsequently, extensive research has been conducted on the involvement of TRIM28 in various facets of cellular biology, resulting in the elucidation of the intricate characteristics of the TRIM28 protein. Notably, one of the primary pathways in which this pivotal gene exerts its influence is cancer. Table 2 shows a list of proteins that interact with or target by TRIM28 in cancers (Figure 1).
| TRIM family | AML | HCC | CRC | Squamous carcinoma | Lung carcinoma | Glioblastoma | Pancreatic carcinoma | Ovarian cancer | Breast carcinoma | NPC | Kidney cancer | Melanoma | Gastric carcinoma | HEC | Testicular cancer | Thyroid cancer | Cervical cancer | Prostate cancer | HNSCC | Osteosarcoma | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRIM1 | * | * | * | 38-40 | |||||||||||||||||
| TRIM2 | * | * | * | * | * | * | * | * | * | * | * | * | 38, 41-44 | ||||||||
| TRIM3 | * | * | * | * | * | * | * | * | * | * | 9, 39, 41, 45-49 | ||||||||||
| TRIM4 | * | * | * | 50-52 | |||||||||||||||||
| TRIM5 | * | * | 43, 53 | ||||||||||||||||||
| TRIM6 | * | * | * | * | 38, 54-56 | ||||||||||||||||
| TRIM7 | * | * | * | * | * | * | * | 9, 43, 57-61 | |||||||||||||
| TRIM8 | * | * | * | * | * | * | * | * | * | * | 39, 41, 62-65 | ||||||||||
| TRIM9 | * | * | * | * | 39, 66, 67 | ||||||||||||||||
| TRIM10 | * | * | * | 9, 68 | |||||||||||||||||
| TRIM11 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 39, 41, 43, 54, 69-77 | ||||||
| TRIM13 | * | * | * | * | * | * | 38, 40, 41, 43, 78 | ||||||||||||||
| TRIM14 | * | * | * | * | * | * | * | * | * | * | * | * | 9, 39, 41, 79-84 | ||||||||
| TRIM15 | * | * | * | * | 39, 41, 85, 86 | ||||||||||||||||
| TRIM16 | * | * | * | * | * | * | * | * | * | * | 41, 54, 87, 88 | ||||||||||
| TRIM17 | * | * | * | * | * | 89, 90 | |||||||||||||||
| TRIM21 | * | * | * | * | * | * | * | * | * | * | * | * | 41, 43, 53, 91-97 | ||||||||
| TRIM22 | * | * | * | * | * | * | * | * | * | * | * | 9, 39, 41, 43, 79, 98-103 | |||||||||
| TRIM23 | * | * | * | 9, 104 | |||||||||||||||||
| TRIM24 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 9, 10, 39, 41, 54, 105-108 | ||||||
| TRIM25 | * | * | * | * | * | * | * | * | * | * | 9, 41, 43, 109, 110 | ||||||||||
| TRIM26 | * | * | * | * | * | * | * | * | 9, 38, 40, 111-115 | ||||||||||||
| TRIM27 | * | * | * | * | * | * | * | * | * | * | * | * | 41, 116-118 | ||||||||
| TRIM28 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 10, 41, 43, 54, 119-123 | |||
| TRIM29 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 9, 39, 41, 86, 124 | ||||
| TRIM31 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 39, 41, 54, 79, 125-128 | ||||
| TRIM32 | * | * | * | * | * | * | * | 9, 41, 53, 80, 129, 130 | |||||||||||||
| TRIM33 | * | * | * | * | * | * | * | * | * | 9, 10, 41, 131-133 | |||||||||||
| TRIM35 | * | * | * | * | 9, 38, 134, 135 | ||||||||||||||||
| TRIM36 | * | * | * | * | * | * | 136-138 | ||||||||||||||
| TRIM37 | * | * | * | * | * | * | * | * | * | * | * | * | 41, 54, 70, 139 | ||||||||
| TRIM38 | * | 140 | |||||||||||||||||||
| TRIM39 | * | * | * | * | 126, 141, 142 | ||||||||||||||||
| TRIM44 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 9, 40, 41, 53, 143-149 | ||||
| TRIM45 | * | * | * | 54, 150, 151 | |||||||||||||||||
| TRIM46 | * | * | * | 152, 153 | |||||||||||||||||
| TRIM47 | * | * | * | * | * | * | * | * | 38, 40, 41, 53, 154, 155 | ||||||||||||
| TRIM48 | * | 156 | |||||||||||||||||||
| TRIM50 | * | * | * | * | 9, 38, 41, 157, 158 | ||||||||||||||||
| TRIM52 | * | * | * | * | 54, 153, 159 | ||||||||||||||||
| TRIM54 | * | 160 | |||||||||||||||||||
| TRIM55 | * | * | * | * | * | 9, 38, 43, 161, 162 | |||||||||||||||
| TRIM56 | * | * | * | * | * | * | * | * | 9, 43, 163 | ||||||||||||
| TRIM58 | * | * | * | * | * | * | * | * | 9, 41, 105, 164-167 | ||||||||||||
| TRIM59 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 41, 54, 168-173 | ||||||
| TRIM62 | * | * | * | * | * | * | * | 9, 41, 68, 174 | |||||||||||||
| TRIM63 | * | * | * | * | 41, 43, 175, 176 | ||||||||||||||||
| TRIM65 | * | * | * | * | * | * | 41, 177-181 | ||||||||||||||
| TRIM66 | * | * | * | * | * | * | * | 10, 41, 54, 169, 182, 183 | |||||||||||||
| TRIM67 | * | * | * | 184 | |||||||||||||||||
| TRIM68 | * | * | 41, 185 | ||||||||||||||||||
| TRIM71 | * | * | * | * | 9, 186-188 | ||||||||||||||||
| TRIM72 | * | * | 53, 189 | ||||||||||||||||||
| TRIM73 | * | * | * | * | 39, 157 |
- * indicates TRIM family members that have been previously reported in the specific cancer types listed.
- Abbreviations: AML, acute myelocytic leukemia; CRC, colorectal cancer; HCC, hepatocellular carcinoma; HEC, human esophageal cancer; HNSCC, head and neck squamous cell carcinoma; NPC, nasopharyngeal carcinoma.
| Modification | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author | Target | Poly-ubiquitination | Deacetylation | HDAC1-mediated deacetylation | SUMOylation | None | Effect | Oncogenic | Tumor suppressive | References |
| Pineda, Carlos T., et al. 2015 | AMPK | * | Degradation of AMPK | + | 190, 191 | |||||
| Pineda, Carlos T. et al. 2015 | ||||||||||
| Venkov, Christo D., et al. 2007 | CBF-A/FTS-1 | * | Upregulation of mesenchymal markers such as VIM and FSP1 | + | 192 | |||||
| Hu, Chen, et al. 2012 | E2F1 | * | The suppression of E2F1 activity | + | 193 | |||||
| Chen, Lu, et al. 2012 | E2F3, E2F4 | * | The deactivation of E2F3 and E2F4 | + | 194 | |||||
| Jin, X., et al. 2017 | FBP1 | * | Degradation of FBP1 | + | 195 | |||||
| Okamoto, Koji, et al. 2006 | P53 | * | * | Inactivation/degradation of P53 | + | 196, 197 | ||||
| Wang, Chuangui, et al. 2005 | ||||||||||
| Wei, Chunli, et al. 2016 | TWIST1 | * | The interaction and stabilization of TWIST1 | + | 198 | |||||
| Mita, Paolo, et al. 2016 | URI–PP2A | * | The removal of phosphate groups from TRIM28, specifically at the Ser824 site, leads to the condensation of chromatin | + | 199 | |||||
| Yang, Yonghua, et al. 2013 | VPS34 (PI3KC3) | * | Activation of VPS34 | + | 200 | |||||
| Zhang, Ren-Yu, et al. 2021 | UBE2S | * | Accelerates cell cycle by ubiquitination of p27 | + | 201 | |||||
| Yang, Yongkang, et al. 2022 | HIF-1 | * | Release paused RNA polymerase II | + | 202 | |||||
| Yao-Jen Chang, et al. 2023 | STAT3 | * | * | * | Activation of STAT3 signaling, inflammation and tumorigenesis | + | 203 | |||
| Kailang Li, et al. 2024 | BRCA1 | * | * | BRCA1 repression and genomic instability and cancer progression | + | 120 | ||||
- Abbreviations: AMPK, AMP-activated protein kinase; BRCA1, breast cancer gene 1; CBF-A, CArG box-binding factor A; E2F1, E2F transcription factor 1; E2F3, E2F transcription factor 3; E2F4, E2F transcription factor 4; FBP1, fructose-bisphosphatase 1; FSP1, fibroblast-specific protein 1; FTS-1, fibroblast transcription site-1; HIF-1, hypoxia-inducible factor 1; PI3KC3, phosphatidylinositol 3-kinase catalytic subunit type 3; PP2A, protein phosphatase 2; Ser824, serine 824; STAT3, signal transducer and activator of transcription 3; TWIST1, Twist family bHLH transcription factor 1; UBE2S, ubiquitin conjugating enzyme E2 S; URI, unconventional prefoldin RPB5 interactor protein; VIM, vimentin; VPS34, vacuolar protein sorting.
- * indicates types of protein modifications that interact with or target by TRIM28 in various cancers.
- + represents types of oncogenic/tumor-suppressive proteins that interact with or target by TRIM28 in various cancers.
TRIM28 not only contributes to the progression of various tumors and worsens their prognosis, as detailed in Table S1, but it also exhibits anticancer properties in certain key cancers, which are presented in Table 3. These contrasting roles highlight the dual nature of TRIM28’s involvement in cancer, emphasizing the complexity of its function and the importance of understanding its context-dependent effects in cancer biology.
| Type of cancer | Function | References |
|---|---|---|
| Prostate cancer | URI uses PP2A phosphatase to control KAP1 phosphorylation and transcriptional repression. | 199 |
| Kidney cancer | TRIM28 facilitates the ubiquitination and degradation of TFE3, hence preventing the growth of cells that cause renal cell carcinoma. | 204 |
| Hepatocellular carcinoma | Hepatocellular carcinoma is prevented from progressing by HDAC6 through the formation of a transcriptional repression complex with TRIM28. | 205 |
| Colorectal cancer | TRIM28 interacts with CARM1 mechanistically, preventing CARM1 from degrading. | 206 |
| Lung cancer | TRIM28 can modulate cell proliferation by mediating interactions between HDAC1 and E2F. | 194 |
- Abbreviations: CARM1; Coactivator-associated arginine methyl transferase 1, E2F; E2 factor, HDAC1; Histone deacetylase 1, HDAC6; Histone deacetylase 6, KAP1; KRAB domain-associated protein 1, PP2A; Protein phosphatase 2A, and TFE3; Transcription factor E3.
3.2 TRIM28 and cancer
Currently, the clinical significance of TRIM28 in numerous diseases remains unclear. Nevertheless, there are documented instances indicating a connection between the level of TRIM28 expression and cancer. It is worth noting that TRIM28 is also active during developmental stages, exhibiting notably high expression levels in ESCs as well as various tumor types, a topic that will be explored in greater detail. For instance, elevated expression of the TRIM28 gene has been linked to metastatic cervical cancer (CC).207 Increased expression of the TRIM28 gene has also been demonstrated in gastric cancer (GC), correlating with unfavorable prognosis 208; Overexpression of the TRIM28 gene has also been identified in the peripheral blood of patients with GC.207 The expression level of TRIM28 was also found to be elevated in ovarian cancer samples, and this elevation was linked to aggressive clinical characteristics.120 Moreover, high expression of TRIM28 served as an independent predictor for ovarian cancer patients.209 TRIM28 is markedly overexpressed in gliomas, and this overexpression is associated with decreased rates of overall and progression-free survival.210 Additionally, TRIM28 holds promise as a prospective biomarker for predicting tumor classification in glioblastoma.211 Moreover, elevated levels of both mRNA and protein expression of TRIM28 were noted in tumor tissues from patients with liver cancer.120 Similarly, examination of tissue samples has demonstrated an escalation in TRIM28 levels throughout the clinical evolution, rising from approximately 40% in situ invasive breast carcinomas to metastasis in lymph nodes. A significant elevation in TRIM28 gene expression has been noted across all four intrinsic subtypes of breast cancer (BC), as well as in BC metastases, in comparison with normal tissue.212, 213 In osteosarcoma, TRIM28 facilitates the SUMOylation of VPS34 by forming a complex with Plasmacytoma variant translocation-1 (PVT-1). This interaction further promotes the ubiquitination and degradation of TSC2, consequently enhancing the self-renewal and stem cell-like characteristics of osteosarcoma cells.122
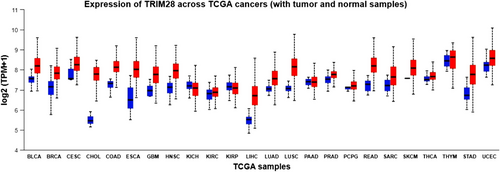
Moreover, there exists a positive correlation between the levels of TRIM28 and the aggressiveness of BC.198 Immunohistochemical analysis has unveiled elevated levels of TRIM28 in a considerable proportion of many cancers, including BC, GC, lung, and so on. This suggests that the upregulation of TRIM28 is a prevalent characteristic among various epithelial cancers.212 To summarize, while numerous studies indicate that increased TRIM28 levels are correlated with poorer prognoses in certain cancers, conflicting findings have also emerged. For example, in early-stage lung cancer, heightened TRIM28 expression is linked to enhanced overall survival, indicating a potential antiproliferative role within tumor cells. Moreover, liver-specific depletion of TRIM28 in mice has been shown to elevate male-predominant hepatic adenoma, indicating a protective role of TRIM28 against tumorigenesis in liver cells.194 Certainly, recent investigations have verified that TRIM28 acts as a crucial regulator of sexual dimorphism in the liver, tightly managing the expression of numerous genes associated with bile and steroid metabolism.214 Hence, TRIM28 plays a crucial role in nontumor cells by ensuring the maintenance of their unaltered phenotype. The majority of cancers (21 out of 24) have elevated TRIM28 expression levels, as seen in Figure 2. But only three cancers—KICH, KIRP, and PAAD—show no discernible difference in TRIM28 expression between normal and cancerous samples. Consequently, it is important to note that TRIM28 affects many types of cancer. The structure and functions of TRIM28, as described, play a significant role in various diseases, particularly in cancer, making its investigation both intriguing and highly relevant. Since studies related to cancer encompass various aspects, the relationship of each aspect with TRIM28 will be thoroughly examined.
3.2.1 TRIM28 and cancer cell proliferation
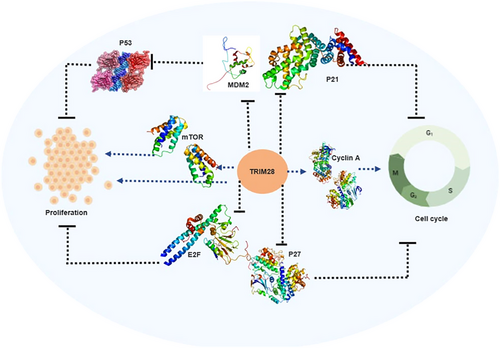
One of the most well-known tumor suppressor genes is TP53, which codes for the P53 protein.215 As an E3 ubiquitin ligase, murine double minute 2 (MDM2) performs a crucial function in the regulation of P53. MDM2 interacts with P53 and facilitates its ubiquitination, representing the primary mechanism governing the levels of P53 in cells.216 In lung cancer, RLIM interacts with MDM2, promoting the ubiquitination-mediated degradation of MDM2. Conversely, TRIM28 binds to RLIM and enhances its ubiquitination, leading to decreased levels of p53 expression. This mechanism ultimately supports tumor cell proliferation and survival.217 Besides its involvement in regulating P53 to influence tumor cell proliferation, TRIM28 can also stimulate CC cell growth by activating the mTOR signaling pathway.218 Likewise, TRIM28 can promote the growth and migration of endometrial cancer cells by activating the AKT/mTOR signaling pathway. This pathway is known for its involvement in regulating cell growth, survival, and migration.219 TRIM28 can additionally facilitate tumor cell proliferation as it is directly targeted by microRNAs. For example, miR-491 is found to be downregulated in glioblastoma multiforme tissues. The reduced expression of miR-491 results in elevated levels of TRIM28, consequently promoting glioma cell proliferation.220 Additionally, miR-140-3p suppresses the proliferation and migration of BC cells by directly targeting and regulating TRIM28 expression.221
In glioma cells, the suppression of TRIM28 results in elevated levels of P21, leading to cell cycle arrest in the G1 phase.210 UBE2S, which is found to be overexpressed in hepatocellular carcinoma (HCC), interacts with TRIM28 in the nucleus, together promoting the ubiquitination of P27. This collaborative mechanism aids in facilitating cell cycle progression.201 TRIM28 can also exhibit an antiproliferative effect in early lung cancer by inhibiting the transcriptional activity of the E2F family.194
3.2.2 The function of TRIM28 in cancer cell apoptosis
The interaction between TRIM28 and MDM2 contributes to the suppression of apoptosis in cancer cells.197 The MAGE protein, frequently upregulated in various cancer types, acts as a corepressor of P53 by forming a complex with TRIM28.222 Reducing TRIM28 levels in non-small cell lung cancer leads to decreased expression of the antiapoptotic gene Bcl-2, while simultaneously increasing the expression of the proapoptotic genes Bax and P53.218 TRIM28 can also negatively regulate the expression of proapoptotic genes, such as BOK, at the posttranscriptional level.223 Activation of RIPK3 leads to the phosphorylation of TRIM28 at S473, causing a disruption in TRIM28’s ability to bind to chromatin.224 In a previous study, the authors have shown that the introduction of TRIM28 siRNA leads to a reduction in cell proliferation and impedes cell cycle progression in NSCLC cell lines.218 The antiapoptotic protein BCL2A1, a member of the BCL-2 family, is implicated in chemoresistance in certain tumors. Despite its significance, BCL2A1 has a brief half-life, largely due to ongoing processing by the ubiquitin–proteasome system, which serves as a crucial tumor-suppressor mechanism governing BCL2A1 activity. However, the specific enzymes responsible for modulating BCL2A1 protein stability remain unknown. Interestingly, endogenous TRIM28 and BCL2A1 have been observed to interact with each other at mitochondria. Knockdown of TRIM28 leads to reduced ubiquitination of BCL2A1.225
3.2.3 TRIM28 and epithelial-to-mesenchymal transition
The process known as epithelial-to-mesenchymal transition (EMT) is characterized by the loss of epithelial traits, notably cell polarity and intercellular connections, coupled with the acquisition of a mesenchymal phenotype.226 The initiation of EMT can be prompted by various extracellular stimuli and transcriptional regulators. However, the precise mechanisms through which these disparate signaling pathways coordinate the intricate cellular processes that enable EMT remain incompletely understood.227, 228 Initially identified during various pivotal phases of embryonic development, EMT has been associated with facilitating carcinoma invasion and metastasis.229 Primary features of EMT include diminished levels of cell adhesion molecules, particularly E-CADHERIN; (ii) EMT entails increased expression of matrix metalloproteinases, facilitating basement membrane degradation; (iii) EMT involves the initiation of the Rho/Cdc42 family small GTPases, crucial for restructuring the cytoskeleton 230; and (iv) during EMT, several transcription factors, including β-CATENIN, the TCF/LEF1 complex, SNAI1, SNAI2, SIP-1, and TWIST1, undergo nuclear translocation.231 Moreover, the presence of specific EMT inducers has been identified in various human cancer specimens, including breast carcinomas.227, 229 A recent breakthrough has revealed a new central controller of EMT, consisting of a protein–DNA compound featuring TRIM28 protein, CBF-A, and the FTS-1 element. This compound holds significant importance in managing the expression of FSP1 in fibroblasts.192
The CBF-A and TRIM28 proteins possess the capability to identify and attach to the FTS-1 sites within genomic DNA, consequently regulating the expression of a wide range of EMT-responsive genes.232 The presence of these proteins at the FTS-1 site within chromatin of epithelial cells undergoing transition to fibroblasts is associated with the activation of the EMT proteome.192 Recent studies suggest that TRIM28 has the ability to facilitate the infiltration of cancer cells by regulating EMT.16 Likewise, it has been noted that heightened levels of TRIM28 prompt EMT in pancreatic cancer cells, both in laboratory settings and within living organisms.233 Furthermore, studies have demonstrated that TRIM28 is implicated in TGF-β-triggered EMT in non-small cell lung cancer cells.234 A recent report has unveiled that TRIM28 fosters BC metastasis by stabilizing the TWIST1 protein, acknowledged as a transcription factor pivotal in serving as a regulator of EMT.198
Both overexpression and depletion of TRIM28 resulted in changes in TWIST1 protein levels, with upregulation and downregulation, respectively, without affecting TWIST1 mRNA levels. Knockdown of TRIM28 caused TWIST1 to be downregulated, accompanied by increased E-CADHERIN expression and decreased N-CADHERIN expression.198 Importantly, EMT plays a pivotal role in obtaining and maintaining stem cell-like traits, capable of imparting stem cell features to both differentiated normal and cancerous cells.235 Moreover, cancer stem cells (CSCs) often exhibit characteristics associated with EMT. The bidirectional interaction between EMT and CSCs carries substantial implications for tumor progression. Considering the known role of TRIM28 protein in governing EMT, there arises the possibility of its involvement in sustaining CSCs. Consequently, TRIM28 emerges as a promising candidate as an epigenetic modulator that promotes cellular metastasis. Therefore, it could represent a promising therapeutic target for addressing cancer, especially when used in conjunction with conventional treatment modalities.196
3.2.4 TRIM28 and CSC attributes
ESCs have been widely employed as a model to explore the molecular mechanisms underlying self-renewal and pluripotency.236 The role of TRIM28 protein in mouse embryonic development was first clarified in 2000.237 TRIM28 plays a critical role in preserving cells in their inherent “state of cell differentiation,” thereby promoting stem cell maintenance while also impeding the reprogramming of somatic cells.238 Observations revealed that the reduction of TRIM28 led to a notable decrease in the mRNA expression levels of Oct-4, Sox2, and Nanog, subsequently triggering the differentiation of ESCs towards the primitive ectoderm lineage.238 Alongside other pluripotency markers like Cnot3, Zfx, and c-Myc, TRIM28 simultaneously binds to numerous potential gene promoters.238 Moreover, investigations have demonstrated that phosphorylated TRIM28 protein at Ser824 interacts with the pluripotency-specific transcription factor Oct-4 on the promoters of genes associated with pluripotency. As a result, this mechanism contributes to the efficient regulation of pluripotent ESCs in a manner dependent on phosphorylation.239 Furthermore, it has been reported that TRIM28 exhibits contrasting effects on differentiation-inducible markers in mouse ESCs, in contrast to its role in inducing the expression of pluripotency markers.240 TRIM28 has been identified as an epigenetic barrier to induced pluripotent stem (iPS) cell reprogramming.241 Interestingly, it has been documented that while depletion of TRIM28 relaxes chromatin and facilitates the reversion to a differentiated state, iPS cells generated with reduced TRIM28 expression swiftly lose their self-renewal potential and undergo spontaneous differentiation. This underscores the crucial role of TRIM28 in sustaining stable iPS cells.242 Recently, the crucial role of TRIM28 in sustaining the population of BC stem cells has been documented.243
CSCs represent a small subset of cells within tumors characterized by their possession of stem cell-like attributes, such as self-renewal capacity, pluripotency, heightened tumorigenic potential, and resistance to therapy.244 The suppression of TRIM28 has been demonstrated in the highly aggressive, undifferentiated cells of the triple-negative BC MDA-MB-231 cell line.212 Additionally, the depletion of TRIM28 led to a confirmed decrease in the population of CSCs in MDA-MB-231 xenografts, as validated by in vivo limiting dilution assays. Furthermore, it was observed that TRIM28, in partnership with EZH2, a component of the PRC2 complex, collectively regulates a subset of genes related to stem cell maintenance and associated with unfavorable survival outcomes in BC patients.243 The depletion of TRIM28 in the MCF7 BC cell line in vitro resulted in a notable decrease in the formation of CD44+ CD24−/low mammospheres, which was accompanied by a reduction in the expression of genes associated with stem cells. These findings underscore the significant role of TRIM28 in activating gene expression that sustains the enrichment and maintenance of mammary stem cells, thereby emphasizing its involvement in promoting cancer progression.238, 243 CD133 serves as a well-studied marker for the most malignant tumor cell population, known as CSCs. However, the precise function of this glycoprotein and its involvement in cellular regulatory pathways remain inadequately understood. Notably, among these pathways, the TRIM28 transcription factor holds particular significance. Experimental evidence, particularly in Caco2 cell line clones, confirms the pivotal role of TRIM28 in modulating CD133 expression. Specifically, knockout of the TRIM28 gene resulted in the downregulation of CD133 expression. These findings underscore the significant contribution of the TRIM28 transcription factor in regulating the heterogeneity of cancer cells associated with CD133.245
3.2.5 TRIM28 and cancer cell metabolism
Cancer cells employ diverse strategies, such as increased glycolytic activity, modulation of redox signaling, and regulation of autophagy, to evade cell death and counteract nutrient deprivation.246 Recent studies have shown that autophagy is mechanistically linked to the preservation of CSCs, allowing them to survive drug toxicity.247, 248 Remarkably, the TRIM28 protein is involved in regulating autophagy at multiple levels.190, 191, 200 There have been reports indicating a notable involvement of the TRIM28 protein in orchestrating phagophore formation.200 The initiation and nucleation of the phagophore rely on the activity of AMP-AMPK and ULK1/2 complex, which is regulated by the mTORC1.249 VPS34 recruits other autophagy regulatory proteins essential for phagophore and autophagic vesicle (AV) formation.250 In conjunction with its binding partner, BECLIN1, VPS34 forms a complex that is additionally associated with the TRIM28 protein. Through its role as a SUMO E3 ligase via its PHD domain, TRIM28 facilitates the SUMOylation of VPS34 at Lys840, consequently augmenting its activity and promoting autophagosome formation.200 Additionally, TRIM28 is implicated in the regulation of mitophagy, a process that involves the selective degradation of mitochondria by autophagy.251 This interaction leads to reduced AMPK signaling, increased mTORC signaling, and subsequent inhibition of autophagy.247, 248 Moreover, it has been shown that the assembly of MAGE–TRIM28 ubiquitin ligase complexes enhances the Warburg effect and facilitates the progression of HCC by directing the degradation of FBP1, a key enzyme in gluconeogenesis.195
3.2.6 TRIM28 and silencing of TEs
Genetic instability is a key characteristic linked to the development and advancement of cancer. TEs are among the factors contributing to this instability, as they can cause various types of cancer by inserting mutations into specific genes that are crucial for either suppressing or promoting malignant transformation.252 Increased levels of TRIM28 have been noted to enhance the utilization of glucose and the generation of lactate by facilitating the degradation of FBP1 in HCC. This action promotes the proliferation of cancer cells both in laboratory settings and in a murine model, indicating a significant role for TRIM28 in promoting tumor growth. These discoveries underscore the protumorigenic properties of TRIM28. Clearly, the interplay between TRIM28 and the metabolic status of cancer cells is intricate and requires additional investigation.253
In human cells, TEs encompass both DNA transposons and RNA transposons, commonly referred to as retroelements. The activity of RNA transposons is meticulously regulated in ESCs through a precisely orchestrated epigenetic mechanism involving KRAB-ZNF repressors in conjunction with their cofactor, TRIM28.254, 255 Ten years ago, TRIM28 was identified as a component of the repressor binding site-binding complex, which plays a role in epigenetically silencing retroviruses in both ESCs and embryonic carcinoma cells.254, 256 Reducing the expression of TRIM28 resulted in the depletion of the repressive chromatin modification H3K9me3 specifically at 5′UTR of IAP genomes. This alteration led to the heightened expression of IAPs.254 The regulation imposed by TRIM28 on retroelement expression is vital for maintaining the transcriptional dynamics during early embryo development. However, the absence of TRIM28 in mouse embryonic fibroblast cells did not result in increased retrotransposon expression.255 This observation suggests that the silencing of retrotransposons plays a minimal role in differentiated cells.255 A recent discovery highlighted the involvement of TRIM28 in the suppression of LINE-1 in mouse fibroblasts. TRIM28 coordinates the compaction of these TEs into transcriptionally inactive heterochromatin, a process instigated by mono-ADP ribosylation facilitated by the SIRT6 protein.257 This modification is crucial for recruiting HP1α and other silencing factors to the target locus. However, the mechanism by which TRIM28 is directed to specific chromatin sites where the L1 retroelement is located remained unclear.258 Similar findings were also observed in human ESCs.259
However, it remains uncertain whether cancer cells operate under similar mechanisms. Unlike normal cells, many human cancers and cancer-derived cell lines exhibit variable, yet generally elevated levels of endogenous full-length L1 mRNA expression, even in the presence of increased TRIM28 activity.260 A novel URI–PP2A–TRIM28 complex has been reported to play a substantial role in regulating L1 expression in prostate cancer cells, representing a recent advancement in our understanding of L1 regulation.199 URI, identified as a transcriptional repressor interacting with RNA polymerase II, has been shown to associate with and facilitate the dephosphorylation of DNA-bound TRIM28–Ser824 by recruiting the PP2A phosphatase. This process leads to the transcriptional suppression of the L1 retroelement.261 Researchers observed a significant elevation in L1 mRNA expression subsequent to TRIM28–Ser824 phosphorylation, accompanied by chromatin decondensation. This phosphorylation was triggered by URI depletion, leading to the prevention of PP2A recruitment to TRIM28 phospho-Ser824.199
3.2.7 Shared pathway in cancers and TRIM28
TRIM28 exhibits a multifaceted role in cancer pathogenesis by regulating diverse signaling pathways implicated in cell proliferation, survival, and genomic stability. Unraveling the precise mechanisms by which TRIM28 influences different cancer types could offer valuable insights for the design of targeted therapeutic approaches (Table 4 and Figure 3). The development of targeted therapeutic approaches may benefit from a better understanding of the precise mechanisms by which TRIM28 influences various cancer types. This might be taking advantage of its regulatory networks to make cancer cells more susceptible to current therapies or creating brand-new inhibitors that precisely block its carcinogenic properties. Moreover, TRIM28’s capacity to engage with the immune system, especially via its function in immune regulation and inflammation, emphasizes the possibility of using it as a therapeutic target in the tumor microenvironment in addition to cancer. Gaining insight into the entire range of TRIM28’s roles in cancer biology may help create more specialized and successful cancer treatments.
| Type of cancer | Signaling pathway | References |
|---|---|---|
| CC | mTOR | 218 |
| BC | EMT | 198 |
| GC | TBK1-IRF1 and TBK1-mTOR | 262 |
| Endometrial cancer | AKT/mTOR | 219 |
| Ovarian carcinoma | Wnt/β-catenin/Shh | 263, 264 |
| Thyroid cancer | Wnt/β-catenin | 265 |
| Colorectal cancer | EMT | 266 |
| Bladder cancer | mTOR | 267 |
| Glioblastoma | Autophagy | 268 |
| B-cell non-Hodgkin lymphoma cells | p21/ promoting the cell cycle progression | 269 |
| Lung cancer | p53 | 217 |
| Non-small cell lung cancer | p53 | 218 |
| Prostate cancer | Androgen receptor | 270 |
- Abbreviations: AKT, serine/threonine kinase; Shh, sonic hedgehog pathway; EMT, epithelial–mesenchymal transition; IRF1, interferon regulatory factor 1; mTOR, mammalian target of rapamycin; TBK1, TANK-binding kinase 1; Wnt, Wingless-related integration site; β-catenin, beta-catenin.
3.2.8 The involvement of TRIM28 in immune modulation
Targeting TRIM28 offers a promising strategy for therapeutic intervention to enhance radiation-induced antitumor immune responses.271 The SETDB1–TRIM28 complex acts as a major inhibitor of antitumor immunity.272 Melanoma exhibiting high level of TRIM28 expression demonstrated a significant downregulation of members of the IRF transcription factor family, including IRF1, IRF2, IRF5, and IRF8, which correlated with diminished IFN signaling.273 Moreover, TRIM28 acts as a specific SUMO E3 ligase for IRF7. It also interacts with the N-terminus of IRF5, thereby inhibiting its function in inflammatory macrophages.274 TRIM28 associates with the transcriptional regulator FOXP3 in human Tregs.275, 276
Notably, TRIM28 knockdown has been associated with increased responsiveness to anti-PD-1 therapy in immunocompetent mice, evidenced by higher levels of CD8+ T tumor-infiltrating lymphocytes and decreased numbers of myeloid-derived suppressor cells.277
The SETDB1–TRIM28 complex has been found to exhibit a negative correlation with the infiltration of effector CD8+ T cells. Inhibiting the SETDB1–TRIM28 complex has been shown to simultaneously upregulate PD-L1 expression and activate the cyclic GMP–AMP synthase cGAS–STING innate immune response pathway, thereby increasing the infiltration of CD8+ T cells. Mechanistically, inhibition of SETDB1–TRIM28 results in the formation of micronuclei in the cytoplasm, which is known to activate the cGAS–STING pathway.272
3.2.9 TRIM28 and autophagy
Autophagy has a multifaceted role in cancer, exhibiting both protumorigenic and antitumorigenic effects that vary with the stage and context of the disease. TRIM proteins, as regulators of autophagy, play a significant role in this dual nature, acting either as proto-oncogenes or antioncogenes. For instance, the MAGEA3 or 6 interacts with TRIM28 to ubiquitinate and degrade PRKAA2, which leads to autophagy inhibition through the mTOR pathway and subsequently promotes tumorigenesis. In the absence of MAGEA3 or 6, TRIM28 instead promotes autophagy by SUMOylating PIK3C3, which facilitates the formation of the BECN1 complex. Additionally, TRIM28 is implicated in the pathogenesis of glioblastoma by inducing autophagy.278, 279 TRIM28 functions as both a proautophagy and antiautophagy factor under different circumstances, reflecting a dichotomy resulting from the coordinated actions of multiple factors. For instance, TRIM28 can inhibit autophagy by associating with MAGEA3 or 6 to ubiquitinate and degrade PRKAA2, thereby activating the mTOR pathway and promoting tumorigenesis. Conversely, in the absence of MAGEA3 or 6, TRIM28 can promote autophagy through the SUMOylation of PIK3C3, which enhances BECN1 complex formation. This dual functionality underscores the complex regulatory role of TRIM28 in autophagy, influenced by its interactions with various molecular partners.268 It has been reported that TRIM28 is involved in the autophagy process. Evidence indicates that TRIM28 activates autophagy and enhances cell proliferation in glioma.268 To put it differently, autophagy assumes different functions at distinct phases of tumor advancement. Specifically, it typically impedes tumor advancement during the initial phases of cancer progression, whereas it fosters tumor expansion in later stages.200 TRIM28 is indispensable for the synthesis of inosine phosphate and the generation of AVs in the process of autophagy.200 TRIM28 also participates in the control of mitophagy, which involves the targeted breakdown of mitochondria via autophagy.251 In gliomas, there is a notable increase in the expression levels of TRIM28 and autophagy as the tumor grade advances. Suppression of TRIM28 can impede autophagy in glioblastoma cells.268 TRIM28 has the capability to degrade AMPK, a cellular energy sensor and regulator, through ubiquitination pathways. This degradation leads to a substantial decrease in autophagy, changes in cellular metabolism, and activation of mTOR signaling.190 The MAGE–TRIM28 axis also influences the Warburg and the advancement of HCC by targeting FBP1 and promoting its degradation. This suggests that the MAGE–TRIM28 axis governs the metabolic reprogramming of cancer cells by regulating the degradation of various metabolic regulators.195 TRIM28 appears to play a crucial role in regulating tumor cell apoptosis, necroptosis, and autophagy. Nonetheless, its association with pyroptosis and ferroptosis remains unexplored. Investigating whether TRIM28 is implicated in these two forms of cell death is essential for a comprehensive understanding of its role in tumor biology.
3.2.10 TRIM28 and necroptosis
Necroptosis is a type of cell death that induces inflammation and, while it shares some morphological similarities with necrosis, it is fundamentally different. Unlike necrosis, necroptosis is a regulated process akin to apoptosis, involving specific death signaling pathways that lead to controlled cell death. The TNF (tumor necrosis factor)/TNFR (TNF receptor) signaling pathway is a well-established trigger for necroptosis. When TNF binds to TNFR1, it activates downstream signaling pathways. Under certain conditions, such as the inhibition or deficiency of caspase-8, TNFR1 signaling can initiate the necroptosis pathway, resulting in necroptotic cell death. This pathway has been thoroughly investigated and serves as a critical model for elucidating the mechanisms and regulation of necroptosis.280, 281 RIPK1, an upstream regulator, is indeed considered a key signaling node in the TNF signal transduction pathway. Depending on the cellular environment and the presence of other signaling molecules, RIPK1 can determine whether TNF stimulation results in cell survival, apoptosis, or necroptosis. RIPK1's role is intricately regulated and essential for maintaining cellular homeostasis and responding to stress signals. When apoptosis is blocked, such as due to inhibited or deficient caspase-8 activity, RIPK1 forms a complex with RIPK3 known as the necrosome. This necrosome complex is crucial for mediating necroptotic cell death, acting downstream of various death receptor signaling pathways, including TNF/TNFR1. The necrosome activates downstream effectors that drive necroptosis, a controlled form of necrotic cell death.282 The necrosome phosphorylates the activation loop of MLKL (mixed-lineage kinase domain-like protein), which is the essential final effector in the necroptotic pathway. Once phosphorylated, MLKL translocates to the plasma membrane, where it compromises membrane integrity. This disruption leads to the release of intracellular contents and triggers inflammation, hallmark features of necroptotic cell death.283-285 Activation of MAPK14 (mitogen-activated protein kinase 14), driven by oligomerized MLKL (mixed lineage kinase domain-like pseudokinase), leads to the phosphorylation of TRIM28 at S473. This phosphorylation promotes inflammation in the later stages of necroptosis, revealing how RIPK1 kinase activation influences inflammatory responses. Necroptosis enhances tumor immunogenicity and is being explored as a target for cancer immunotherapy due to its ability to induce inflammatory protein synthesis and stimulate antitumor immune responses. TRIM28, a corepressor in necroptosis, is phosphorylated by RIPK3, reducing its chromatin-binding ability and thereby increasing the activation of NFKB and other transcription factors.224
3.2.11 The potential of TRIM28 as a target for cancer therapy
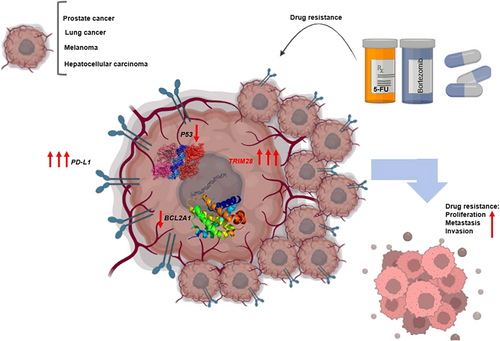
Cytotoxic chemotherapeutic agents and targeted therapies are pivotal in clinical oncology. Nevertheless, the development of drug resistance in tumor cells significantly compromises the efficacy of these anticancer treatments.286 Reducing TRIM28 expression significantly increases the vulnerability of lung cancer cells to 5-fluorouracil.287 Bortezomib (BTZ), a selective inhibitor of proteasomes, shows potential in HCC therapy. Nonetheless, drug resistance poses a considerable challenge to its effectiveness. Remarkably, TRIM28 diminishes BTZ sensitivity in HCC cells by enhancing proteasome expression.288 The expression of TRIM28 is notably elevated in castration-resistant prostate cancers. TRIM28 overexpression amplifies androgen receptor signaling, representing a pivotal mechanism in antiandrogen deprivation therapy for prostate cancer.270 E2F1 is a frequently targeted molecule in the development of anticancer chemotherapeutics.289 This phenomenon may be linked to the expression of TRIM28, which recruits HDAC1 to deacetylate E2F1, thereby suppressing its activity. Additionally, the downregulation of TRIM28 enhances cell death induced by etoposide.33 Following DNA damage, phosphorylation of TRIM28 at Ser473 boosts its interaction with E2F1. Utilizing combination chemotherapy involving etoposide along with a suitable TRIM28 S473p-blocking peptide might aid in inducing tumor cell death. Additionally, the P53 pathway represents a significant focus in cancer drug development.290
In cells where TRIM28 is depleted, exposure to actinomycin D results in the activation of P53, suggesting that TRIM28 could serve as a target for P53 reactivation in cancer cells. Coadministration of actinomycin D with compounds that disrupt the interaction between TRIM28 and MDM2 or decrease TRIM28 levels can effectively inhibit MDM2 function, thereby significantly activating P53.196, 291 The peroxide-induced activation of P38 MAPK promotes the phosphorylation of TRIM28 at S473, resulting in the activation of TRIM28. This activated state effectively assists DNA repair mechanisms, thereby aiding tumor cells in surviving exposure to exogenous ROS.292 Hence, inhibiting TRIM28 may render tumor cells more susceptible to chemotherapy.
A wide range of nanobodies has been utilized in clinical trials aimed at treating various diseases across the spectrum.293 The specific anti-TRIM28 nanobody, NB237, has been demonstrated to significantly inhibit the invasion and metastasis of both glioblastoma cells and glioblastoma stem cells within the zebrafish brain.294 Research conducted in cardiomyocytes has demonstrated that ZFG protects against hypoxia/reoxygenation-induced apoptosis by decreasing the expression of TRIM28.295 There is an urgent need for the development of small molecule inhibitors targeting TRIM28, which could potentially serve as adjuncts to tumor therapy.267 PD-L1 plays a crucial role in facilitating tumor-induced immunosuppression,296 and verteporfin has emerged as a significant small molecule inhibitor of PD-L1. Studies suggest that verteporfin can inhibit both basal and IFN-induced PD-L1 expression, both in laboratory settings and within living organisms, by targeting Golgi-associated autophagy and disrupting the STAT1–IRF1–TRIM28 signaling cascade.297 Collectively, TRIM28 appears to contribute to drug resistance in certain cancers and is a significant factor rendering certain anticancer drug targets unresponsive to treatment. Advancing tumor treatment goals may involve the development of drugs or small molecules specifically targeting TRIM28. Table 5 provides a list of some cancers related to TRIM28 treatment. This aspect remains a gap in current TRIM28 research efforts (Figure 4).
| Cancer | Clinical evidence | References |
|---|---|---|
| Lung | Reduction of TRIM28 expression enhances the sensitivity to 5-FU, etoposide, and cisplatin. | 298 |
| TRIM28 can upregulate miR-125b-5p and plays a significant role in DDP resistance in patients. | 299 | |
| Knockdown of TRIM28 triggers cell apoptosis by promoting E2F1 inactivation and decreasing cellular sensitivity to etoposide treatment. | 300 | |
| HCC | Reduces sensitivity to BTZ by upregulating proteasome expression. | 288 |
| TRIM28 can increase sensitivity to oxaliplatin therapy by facilitating the ubiquitin-mediated degradation of HMGB1. | 301 | |
| Elevated TRIM28 expression contributes to resistance against TKI therapy. | 15 | |
| Pancreatic | Overexpression of TRIM28 results in enhanced degradation of FBP1, which subsequently prevents the degradation of c-Myc, thereby increasing cellular resistance to bromodomain inhibitor. | 302 |
| Glioma | ATM inhibitor (ATMi) drugs enhance cellular sensitivity to radiotherapy by blocking TRIM28 phosphorylation. | 303 |
| Various cancers | Verteporfin may inhibit PD-L1 expression by disrupting the interaction between TRIM28 and IRF1. | 297 |
- Abbreviations: 5-FU, 5-fluorouracil; BTZ, bortezomib; c-Myc, cellular myelocytomatosis oncogene; DDP, cisplatin; FBP1, fructose-bisphosphatase 1; HMGB1, high mobility group box 1; IRF1, interferon regulatory factor 1; PD-L1, programmed cell death 1 ligands; TKI, tyrosine kinase inhibitors.
3.3 TRIM28 and infertility
Haploinsufficiency, a process where a loss-of-function mutation results in only a single functional copy of a gene, leading to an inadequate production of the gene's product, is a significant contributor to various developmental disorders, including male infertility.304 Phenotypes associated with haploinsufficiency usually involve critical genes that encode transcription factors or are involved in developmental pathways.305-308 It is not surprising that numerous epigenetic factors involved in development, such as NSD1, SUZ12, TRIM28, and others, have been found to exhibit haploinsufficiency phenotypes.309 TRIM28 serves as a crucial mediator of epigenetic modifications and is essential for development, as demonstrated by the embryonic lethality observed in both zygotic and maternal TRIM28 knockout mice, and the rapid mortality in adult mice following systemic TRIM28 deletion (Table 6). Functionally, TRIM28 is involved in a wide range of biological processes, including cell differentiation, the DNA damage response, suppression of retroviral elements, and the epigenetic inheritance from germline to soma.7, 237, 310-312 TRIM28 haploinsufficiency has been demonstrated to induce a bistable obesity phenotype in mice, which is linked to the dysregulation of nonclassically imprinted genes.313 One report reveals a new aspect of TRIM28 haploinsufficiency: a highly penetrant infertility phenotype observed in male mice heterozygous for TRIM28 (TRIM28Het). Currently, TRIM28 is known to be expressed in spermatocytes and round spermatids and is essential for normal spermatogenesis. However, the underlying mechanism of testicular degeneration following TRIM28 deletion remains unclear, with a potentially paracrine, noncell-autonomous mechanism being suggested.314, 315 Premature ovarian insufficiency (POI) is a clinical condition characterized by ovarian dysfunction and abnormal alterations in hormone levels, such as follicle-stimulating hormone and estradiol (E2). Research has shown that POI is associated with the cellular senescence of ovarian granulosa cells, with TRIM28 playing a pivotal role in regulating oxidative stress (OS)-induced cellular senescence in these cells. Mechanistically, OS decreases TRIM28 protein levels in KGN cells, leading to increased expression of autophagy markers ATG5 and LC3B-II, alongside the downregulation of P62.316
| Description | Animal | Disorder | References |
|---|---|---|---|
| TRIM28 likely functions as an E3 SUMO ligase, affecting the stability and subcellular localization of α-Syn and tau via SUMOylation. | Mice | Parkinson's disease and Alzheimer's disease | 317 |
| TRIM28, a crucial epigenetic regulator, undergoes progressive testicular degeneration. | Mice | Infertility | 315 |
| TRIM28 haploinsufficiency results in a stochastic bi-stable phenotype, characterized by the development of obesity or an alternative, nonobese state, also known as polyphenism. | Mice | Epigenetic obesity | 313 |
| Ectopic expression of TRIM28 promoted tumor growth, elevated PD-L1 expression, and inhibited T cell activation. | Mice | GC | 262 |
| TRIM28 protein modulates the tumor growth of the CSC population. | Mice | BC | 212 |
| TRIM28 was highly enriched in the core of the tumor and correlated with the expression of stem cell-related genes. | Zebrafish | Glioblastoma | 294 |
| The deletion of TRIM28 in skeletal muscle of mice, either during development (MCK–cre) or after development (ACTA1–cre–ERT2), does not confer protection against high-fat diet (HFD)-induced obesity or glucose intolerance | Mice | Diabetes | 318 |
- Abbreviations: ACTA1, actin alpha 1; ERT2, estrogen receptor T2; HFD, high-fat diet; MCK, muscle creatine kinase.
3.4 TRIM28 and obesity
The effective storage of lipids in white adipose tissue (WAT) is essential for maintaining systemic energy homeostasis. Numerous genes have been implicated in the regulation of WAT lipid metabolism, including TRIM28, which has traditionally been associated with influencing adiposity through epigenetic mechanisms during embryonic development. However, the present study reveals that mice with adipocyte-specific TRIM28 deletion also exhibit obesity, similar to the phenotype seen in models with global TRIM28 deletion, suggesting a role for TRIM28 beyond its developmental functions (Table 6). Notably, the obesity phenotype was significantly more evident in female mice, suggesting that TRIM28 acts as a sex-specific regulator of obesity. Mechanistically, this phenotype is associated with changes in lipolysis and triglyceride metabolism, which can be attributed in part to the downregulation of Klf14—a gene recognized for its role in modulating adipocyte size and body composition in a sex-specific context. These findings position TRIM28 as a crucial sex-specific regulator of adiposity after developmental and the function of WAT.319
3.5 TRIM28 and diabetes
Diabetes mellitus (T2DM), characterized by insulin resistance and mitochondrial dysfunction in skeletal muscle, is a major cause of mortality in developed countries. There is considerable interest in targeting mitochondrial health, including autophagy pathways, as potential strategies for preventing or treating T2DM. In one study, two distinct muscle-specific TRIM28 knockout models (MCK–cre and ACTA1–cre–ERT2) were examined under both chow and high-fat diet (HFD) conditions (Table 6). Despite muscle-specific TRIM28 deletion leading to alterations in markers of mitochondrial activity and autophagy in skeletal muscle, most metabolic parameters were largely unaffected. In particular, the absence of TRIM28 in skeletal muscle, whether during development (MCK–cre) or postdevelopment (ACTA1–cre–ERT2), did not protect against HFD-induced obesity or glucose intolerance.318 These findings are consistent with prior research on autophagy and mitochondrial function in other cell types, indicating that the role of TRIM28 in regulating mitochondrial function requires further investigation for a more comprehensive understanding.
3.6 TRIM28 and COVID-19
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), utilizes angiotensin-converting enzyme 2 (ACE2) as a receptor to enter human cells.320 The expression level of ACE2 may impact both the susceptibility to and severity of COVID-19, underscoring the importance of understanding the regulatory mechanisms that control ACE2 expression. TRIM28, known for its roles in antiviral defense, maintaining endogenous retrovirus latency, and modulating immune responses, has recently been identified as coexpressed with the SARS-CoV-2 receptor in type II pneumocytes. However, its role in regulating ACE2 expression and facilitating SARS-CoV-2 cell entry into cells remains unclear. The study demonstrated that TRIM28 knockdown resulted in increased ACE2 expression and enhanced pseudotyped SARS-CoV-2 entry into A549 cells and primary pulmonary alveolar epithelial cells.321
3.7 TRIM28 and other viral diseases
TRIM28 plays a major role in cellular defense against viral infections. The immune system, cellular homeostasis, and the body's antiviral responses all depend on it. As an adaptable regulator, TRIM28 affects the host immune system's modulation as well as the direct suppression of viral gene expression.322 Beyond only reacting to active infections, TRIM28 plays a critical role in keeping some viruses in their latent state, which stops reactivation and the subsequent spread of disease. TRIM28’s significance in maintaining the equilibrium of the host's defense mechanisms is further demonstrated by its role in immune regulation. It adjusts inflammatory responses by regulating not only the expression of IFN-stimulated genes but also important signaling pathways like NF-ĸB. This regulatory ability guarantees a sufficiently strong immune response. Next, we discuss the role of TRIM28 in other viral diseases.
The HIV-1 Tat protein, which is necessary for the transcription of the HIV-1 genome, interacts with TRIM28. This interaction promotes viral replication. Moreover, TRIM28 can affect latency and reactivation phases of the HIV life cycle. Histone deacetylases (HDACs) and histone methyltransferases are among the chromatin-modifying enzymes that TRIM28 enlists as transcriptional repressors. By inhibiting the expression of viral genes, these enzymes help to form repressive chromatin structures surrounding the proviral DNA, which keeps HIV latent and transcriptionally inactive.323 As a result, targeting the TRIM28–Tat interaction could offer a novel approach to modulating the transcriptional activity of HIV-1, providing a new avenue for antiviral drug development.
TRIM28 plays a significant role in the regulation of human papillomavirus (HPV) transcription. It can function as an E2F corepressor, which is a crucial transcription factor that HPV targets. This interaction may influence the viral life cycle, including viral replication and persistence, as well as the progression of HPV-related cancers. HPV infection is strongly associated with the development of various cancers, particularly CC.324 The virus expresses oncogenes, such as E6 and E7, which interfere with tumor suppressor proteins like p53 and retinoblastoma, driving the progression toward malignancy.324 TRIM28, by regulating transcription factors and chromatin structure, may influence the expression of these oncogenes. Its role in forming repressive chromatin structures could potentially suppress the expression of viral genes involved in oncogenesis, thereby impacting cancer progression.325 Hence, understanding TRIM28’s role in these processes could provide insights into novel therapeutic strategies for treating HPV-associated diseases.
Hepatitis B virus X protein (HBX) is a multifunctional viral protein and plays an important role in HBV replication and pathogenesis. Through its role as a transcriptional repressor, TRIM28 can affect the transcriptional regulation of viral host genes and HBV replication and interacts with the HBX protein. This interaction has a significant impact on various aspects of the HBV life cycle, including viral replication, transcription, and persistence.326 The interaction between TRIM28 and HBX is particularly important in the context of HBV-induced liver diseases, including chronic hepatitis, liver cirrhosis, and HCC. Therefore, the imbalance between this interaction may affect the development of these diseases. In addition, TRIM28 is involved in the regulation of immune-related genes, and its interaction with HBX may help the virus to evade the host's immune system, leading to chronic infection and an increased risk of liver cancer.326 Therefore, molecular understanding of this interaction can reveal new targets for therapeutic intervention and provide potential strategies to control HBV replication, reduce liver disease progression, and prevent HCC development.
TRIM28 plays an important role in regulating various stages of hepatitis C virus (HCV) replication and assembly. It can modulate the efficiency of viral RNA synthesis and help the virus to infect cells. In addition, chronic HCV infection is one of the main causes of liver diseases, where TRIM28 modulates viral replication and particle aggregation, may contribute to the chronicity of infection at these critical stages.327 Therefore, TRIM28 can be considered as a potential therapy to manage HCV infection and prevent its progression to chronic liver diseases.
Viral infections are intricately linked to the development of tumors. Herpesviruses possess the capability to endure within infected hosts for extended periods without generating viral particles.274 TRIM28 assumes a significant role in tumors associated with herpesviruses, including Epstein–Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), and human cytomegalovirus.328-330 TRIM28 has the capability to sustain the latent phase of EBV and KSHV by binding to the promoters of viral genes.328 TRIM28 becomes a target of latent membrane protein-1 (LMP1)-induced sumoylation, thereby facilitating the maintenance of EBV latency.331 According to recent research, TRIM28 has been found to interact with IFI16, leading to the suppression of EBV lytic gene expression.332 Significantly, the phosphorylation of TRIM28 impedes its suppressive role on virus latency, consequently promoting virus production.333 Moreover, TRIM28 acts as a repressor of human endogenous retroviruses.334 Activation of endogenous retroviruses represents a crucial mechanism in the context of antitumor immune responses triggered by radiotherapy.
4 CONCLUSION AND PROSPECTS
In conclusion, TRIM28, as a key member of the TRIM protein family, plays a pivotal role in maintaining health and contributing to the onset and progression of various diseases. The TRIM family of proteins, particularly TRIM28, has emerged as a pivotal regulator in the pathogenesis of cancer, influencing a multitude of cellular processes that are critical to tumor development and progression. TRIM28 is a key member of the TIF1 family and is characterized by its diverse functional domains, including a RING domain, B-box motifs, and a coiled-coil region, which collectively contribute to its versatility in cellular function. These domains enable TRIM28 to act as a transcriptional coregulator, particularly through its interactions with KRAB-ZNF proteins, thereby playing a significant role in chromatin remodeling and gene expression modulation. Additionally, TRIM28 is deeply involved in the DNA damage response, where its phosphorylation is crucial for the activation and coordination of DNA repair mechanisms. This multifaceted role of TRIM28 underscores its importance not only in maintaining cellular homeostasis but also in the aberrant processes that lead to oncogenesis. Its involvement in various pathways, including those regulating cell cycle progression, apoptosis, and response to genotoxic stress, makes TRIM28 a critical player in cancer biology and a potential target for therapeutic intervention. Understanding the complex regulatory functions of TRIM28 in cancer could provide valuable insights into novel strategies for cancer treatment.
TRIM28’s involvement in transcriptional coregulation, particularly with KRAB-ZNF proteins, underscores its central role in the intricate processes of chromatin remodeling and gene expression modulation. This partnership is crucial for maintaining chromatin structure, ensuring that gene activity is tightly regulated and responsive to the cellular environment. The KRAB-ZNF proteins, a large family of transcriptional repressors, rely on TRIM28 to recruit additional factors necessary for the establishment of repressive chromatin marks. This interaction not only contributes to the silencing of specific genomic regions but also plays a significant role in preserving genome stability. TRIM28 to act as a mediator between chromatin dynamics and DNA repair. The dual role of TRIM28 in gene regulation and DNA repair highlights its critical function in cellular homeostasis and its potential as a therapeutic target in cancer.
In the context of cancer, TRIM28 displays a highly nuanced and context-dependent role, characterized by its ability to function either as a protumorigenic factor or as a tumor-suppressive agent. This duality is influenced by the specific type of cancer and the cellular environment in which it operates. As a protumorigenic factor, TRIM28 contributes to tumor progression by supporting the maintenance of stem cell properties and promoting cellular characteristics conducive to cancer growth. It influences key aspects of tumor biology, including the regulation of the cancer cell cycle, promotion of cell proliferation, and resistance to apoptotic signals, highlighting its significant impact on cancer development.
Conversely, TRIM28 can also exhibit tumor-suppressive properties under certain conditions. Its involvement in the EMT—a process that facilitates cancer metastasis—demonstrates its role in enabling cancer cells to acquire invasive capabilities. Additionally, TRIM28’s functions in maintaining stem cell attributes and silencing TEs further illustrate its multifaceted contribution to cancer progression. These roles collectively emphasize the complexity of TRIM28’s impact on tumor biology and its potential as a target for therapeutic interventions.
TRIM28 influences processes such as neurodevelopment, immune regulation, and metabolic control. Dysregulation of TRIM28 has been linked to a wide range of disorders, including neurodegenerative diseases, autoimmune conditions, and metabolic disorders such as obesity and type 2 diabetes. Its involvement in chronic inflammatory diseases conditions further highlights the importance of TRIM28 in maintaining genomic stability and cellular function. Additionally, TRIM28 plays a significant role in viral infections by modulating the host's immune response and influencing viral replication. This antiviral function underscores TRIM28’s broader role in host defense mechanisms, making it a critical player not only in disease pathogenesis but also in the body's response to viral challenges.
Given its prominent role in diverse aspects of cancer and other disease development, TRIM28 represents a promising candidate for targeted therapeutic strategies. Future research should focus on elucidating the precise mechanisms through which TRIM28 influences disease progression and developing approaches to modulate its activity. By deepening our understanding of TRIM28’s functions and interactions, we can potentially uncover innovative treatment options that harness its capabilities to combat cancer more effectively and enhance patient outcomes.
AUTHOR CONTRIBUTIONS
Formal analysis, review, design of survey and study, writing, and editing: Mazaher Maghsoudloo. Review, formal analysis, writing, and editing: Khatere Mokhtari. Formal analysis and review: Behdokht jamali and Amir Gholamzad. Survey and design, theory development, supervision, project administration, writing of original draft, reviewing and editing of final manuscript: Maliheh Entezari and Mehrdad Hashemi. Concept of the idea, survey and design, theory development, supervision, project administration, writing of original draft, reviewing and editing of final manuscript: Junjiang Fu. All authors read and approved the contents of the manuscript.
ACKNOWLEDGMENTS
We would like to thank the people from the Key Laboratory of Epigenetics and Oncology, the Research Center for Preclinical Medicine, Southwest Medical University. This work was funded by the National Natural Science Foundation of China (grant nos. 82073263 and 81672887) and by Research start-up fund by Foundation of Southwest Medical University (grant no. 41/00170067).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Not applicable.
Open Research
DATA AVAILABILITY STATEMENTETHICS APPROVAL
Not applicable.




