Investigation of genomic DNA methylation by ultraviolet resonant Raman spectroscopy
Corresponding Author
Francesco D'Amico
Elettra-Sincrotrone Trieste, Trieste, Italy
Correspondence
Francesco D'Amico, Elettra-Sincrotrone Trieste S.C.p.A. Strada Statale 14 - km 163,5 in AREA Science Park, 34149 Basovizza, Trieste, ITALY.
Email: [email protected]
Search for more papers by this authorPaolo Zucchiatti
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Physics, University of Trieste, Trieste, Italy
Plasmon Nanotechnologies line, IIT, Genoa, Italy
Search for more papers by this authorKatia Latella
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Chemistry and Industrial Chemistry, University of Genova, Genoa, Italy
Search for more papers by this authorMaria Pachetti
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Physics, University of Trieste, Trieste, Italy
Search for more papers by this authorClaudio Masciovecchio
Elettra-Sincrotrone Trieste, Trieste, Italy
Search for more papers by this authorLorella Pascolo
Institute for Maternal and Child Health, IRCCS Burlo Garofolo, Trieste, Italy
Search for more papers by this authorCorresponding Author
Francesco D'Amico
Elettra-Sincrotrone Trieste, Trieste, Italy
Correspondence
Francesco D'Amico, Elettra-Sincrotrone Trieste S.C.p.A. Strada Statale 14 - km 163,5 in AREA Science Park, 34149 Basovizza, Trieste, ITALY.
Email: [email protected]
Search for more papers by this authorPaolo Zucchiatti
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Physics, University of Trieste, Trieste, Italy
Plasmon Nanotechnologies line, IIT, Genoa, Italy
Search for more papers by this authorKatia Latella
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Chemistry and Industrial Chemistry, University of Genova, Genoa, Italy
Search for more papers by this authorMaria Pachetti
Elettra-Sincrotrone Trieste, Trieste, Italy
Department of Physics, University of Trieste, Trieste, Italy
Search for more papers by this authorClaudio Masciovecchio
Elettra-Sincrotrone Trieste, Trieste, Italy
Search for more papers by this authorLorella Pascolo
Institute for Maternal and Child Health, IRCCS Burlo Garofolo, Trieste, Italy
Search for more papers by this authorAbstract
Cytosine plays a preeminent role in DNA methylation, an epigenetic mechanism that regulates gene expression, the misregulation of which can lead to severe diseases. Several methods are nowadays employed for assessing the global DNA methylation levels, but none of them combines simplicity, high sensitivity, and low operating costs to be translated into clinical applications. Ultraviolet (UV) resonant Raman measurements at excitation wavelengths of 272 nm, 260 nm, 250 nm, and 228 nm have been carried out on isolated deoxynucleoside triphosphates (dNTPs), on a dNTP mixture as well as on genomic DNA (gDNA) samples, commercial from salmon sperm and non-commercial from B16 murine melanoma cell line. The 228 nm excitation wavelength was identified as the most suitable energy for enhancing cytosine signals over the other DNA bases. The UV Raman measurements performed at this excitation wavelength on hyper-methylated and hypo-methylated DNA from Jurkat leukemic T-cell line have revealed significant spectral differences with respect to gDNA isolated from salmon sperm and mouse melanoma B16 cells. This demonstrates how the proper choice of the excitation wavelength, combined with optimized extraction protocols, makes UV Raman spectroscopy a suitable technique for highlighting the chemical modifications undergone by cytosine nucleotides in gDNA upon hyper- and hypo-methylation events.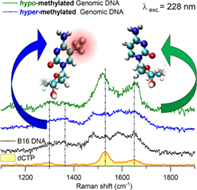
CONFLICTS OF INTEREST
The authors declare no financial or commercial conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data available on request from the authors
Supporting Information
| Filename | Description |
|---|---|
| jbio202000150-sup-0001-supinfo.pdfPDF document, 717.7 KB | Appendix S1: Supporting information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1S. Sharma, T. K. Kelly, P. A. Jones, Carcinogenesis 2010, 31, 27.
- 2G. Felsenfeld, Cold Spring Harb. Perspect. Biol. 2014, 6, a018200.
- 3B. Chowdhury, I. H. Cho, J. Irudayaraj, J. Biol. Eng. 2017, 11, 10.
- 4M. A. Dawson, T. Kouzarides, Cell 2012, 150, 12.
- 5S. Gunes, M. A. Arslan, G. Neslihan, T. Hekim, R. Asci, J. Assist. Reprod. Gen. 2016, 33, 553.
- 6D. Montjean, A. Zini, C. Ravel, S. Belloc, A. Dalleac, H. Copin, P. Boyer, K. McElreavey, M. Benkhalifa, Andrology 2015, 3, 235.
- 7S. Kurdyukov, M. Bullock, Biology 2016, 5, 3.
- 8N. Islam, S. Yadav, H. Haque, A. Munaz, F. Islam, S. Al Hossain, V. Gopalan, A. K. Lam, N. T. Nguyen, M. J. A. Shiddiky, Biosens. Bioelectron. 2017, 92, 668.
- 9J. G. Kelly, G. M. Najand, F. L. Martin, Biophotonics 2011, 4, 345.
- 10M. Banyay, A. Gräslund, J. Mol. Biol. 2002, 24, 667.
- 11L. Li, S. F. Lim, A. Puretzky, R. Riehn, H. D. Hallen, Biophys. J. 2018, 114, 2498.
- 12K. C. Bantz, A. F. Meyer, N. J. Wittenberg, H. Im, Ö. Kurtuluş, S. H. Lee, N. C. Lindquist, S. H. Oh, C. L. Haynes, Phys. Chem. Chem. Phys. 2011, 13, 11551.
- 13J. Morla-Folch, R. A. Alvarez-Puebla, L. Guerrini, J. Phys. Chem. Lett. 2016, 7, 3037.
- 14S. R. Panikkanvalappil, M. A. Mahmoud, M. A. Mackey, M. A. El-Sayed, ACS Nano. 2013, 7(7), 7524.
- 15S. Ganesh, K. Venkatakrishnan, B. Tan, Nat. Comm. 2020, 11, 1135.
- 16S. M. A. Hasan, Y. He, T.-W. Chang, J. Wang, M. R. Gartia, J. Phys. Chem. C 2019, 123, 698.
- 17N. E. Marotta, K. R. Beavers, L. A. Bottomley, Anal. Chem. 2013, 85, 1440.
- 18X. Liu, Y. Shao, Y. Tang, K. F. Yao, Sci. Rep. 2014, 4, 5835.
- 19T. G. Burova, V. V. Ermolenkov, G. N. Ten, D. M. Kadrov, M. N. Nurlygaianova, V. I. Baranov, I. K. Lednev, J. Phys. Chem. A 2013, 117, 12734.
- 20T. G. Burova, V. V. Ermolenkov, G. N. Ten, R. S. Shcherbakov, V. I. Baranov, I. K. Lednev, J. Phys. Chem. A 2011, 115, 10600.
- 21B. E. Billinghurst, S. A. Oladepo, G. R. Loppnow, J. Phys. Chem. B 2009, 113, 7392.
- 22T. Bourova, G. Ten, S. Andreeva, V. Berezin, J. Raman Spectrosc. 2000, 31, 827.
- 23M. Majoube, P. Milliè, L. Chinsky, P. Y. Turpin, G. Vergoten, J. Mol. Struct. 1995, 355, 147.
- 24S. Sasidharanpillai, G. R. Loppnow, J. Phys. Chem. B 2019, 123, 3898.
- 25B. E. Billinghurst, S. A. Oladepo, G. R. Loppnow, J. Phys. Chem. B 2012, 116, 10496.
- 26W. Suen, T. G. Spiro, L. C. Sowers, J. R. Fresco, Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 4500.
- 27A. Toyama, N. Hanada, J. Ono, E. Yoshimitsu, H. Takeuchi, J. Raman Spectrosc. 1999, 30, 623.
- 28S. P. A. Fodor, R. P. Rava, T. R. Hays, T. G. Spiro, J. Am. Chem. Soc. 1985, 107, 1520.
- 29J. Geng, M. Aioub, M. A. El-Sayed, B. A. Barry, ChemPhysChem 2018, 19, 1428.
- 30J. Geng, M. Aioub, M. A. El-Sayed, B. A. Barry, J. Phys. Chem. B 2017, 121, 8975.
- 31J. M. Benevides, S. A. Overman, G. J. Thomas Jr., J. Raman Spectrosc. 2005, 36, 279.
- 32S. P. A. Fodor, T. G. Spiro, J. Am. Chem. Soc. 1986, 108, 3198.
- 33Z. Q. Wen, G. J. Thomas Jr., Biopolymers 1998, 45, 247.
10.1002/(SICI)1097-0282(199803)45:3<247::AID-BIP7>3.0.CO;2-R CAS PubMed Web of Science® Google Scholar
- 34L. Chinsky, P. Y. Turpin, Nucleic Acids Res. 1978, 5, 2969.
- 35P. Zucchiatti, K. Latella, G. Birarda, L. Vaccari, B. Rossi, A. Gessini, C. Masciovecchio, F. D'Amico, J. Raman Spectr. 2018, 49, 1056.
- 36F. Cammisuli, L. Pascolo, M. Morgutti, A. Gessini, C. Masciovecchio, F. D'Amico, Appl. Spectr. 2017, 71, 152.
- 37F. D'Amico, F. Cammisuli, R. Addobbati, C. Rizzardi, A. Gessini, C. Masciovecchio, B. Rossi, L. Pascolo, Analyst 2015, 140, 1477.
- 38J. Marmur, P. Doty, J. Mol. Biol. 1962, 5, 109.
- 39F. D'Amico, M. Saito, F. Bencivenga, M. Marsi, A. Gessini, G. Camisasca, E. Principi, R. Cucini, S. Di Fonzo, A. Battistoni, E. Giangrisostomi, C. Masciovecchio, Nucl. Instrum. Methods Phys. Res., Sect. A 2013, 703, 33.
- 40R. L. McCreery, Photometric Standards for Raman Spectroscopy. in Handbook of Vibrational Spectroscopy, 2006, Vol. 1 (Eds: J. M. Chalmers, R. Peter), John Wiley & Sons Ltd, Chichester (England).
10.1002/0470027320.s0706 Google Scholar
- 41M. J. Cavaluzzi, P. N. Borer, Nucleic Acids Res. 2004, 31, e13.
- 42S. K. Patra, S. Bettuzzi, Biochem. 2009, 74, 613.
- 43C. C. Oakes, S. La Salle, D. J. Smiraglia, B. Robaire, J. M. Trasler, Dev. Biol. 2007, 307, 368.
- 44J. F. Linnekamp, R. Butter, R. Spijker, J. P. Medema, H. W. M. van Laarhoven, Cancer Treat. Rev. 2017, 54, 10.
- 45L. Pascolo, D. E. Bedolla, L. Vaccari, I. Venturin, F. Cammisuli, A. Gianoncelli, E. Mitri, E. Giolo, S. Luppi, M. Martinelli, M. Zweyer, G. Ricci, Reprod. Toxicol. 2016, 61, 39.




