Endothelial single-cell sequencing: A new way to understand endothelial biomedicine
Vascular endothelial cells (ECs) are critical guardians of vascular homeostasis, regulating angiogenesis, inflammation, and barrier integrity. However, their phenotypic and functional heterogeneity across vascular beds has posed challenges to traditional bulk analysis methods. Single-cell RNA sequencing (scRNA-seq) has emerged as a transformative tool, offering unparalleled resolution at the individual cell level. This technology has revolutionized our ability to dissect endothelial diversity and function and unveiled novel endothelial subtypes, unexpected signaling pathways, and dynamic responses to environmental stimuli (Figure 1). scRNA-seq now stands at the forefront of endothelial biology research, providing insights into both physiological and pathological processes.
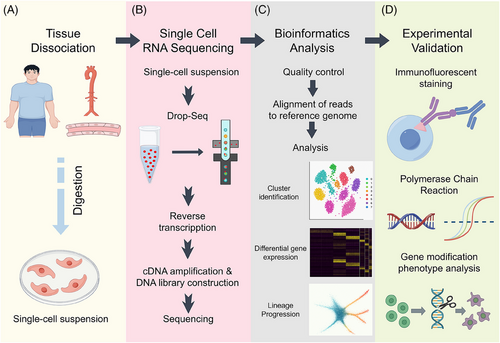
In vascular inflammation, scRNA-seq has transformed our ability to dissect endothelial plasticity and pathological transitions. For instance, McQueen et al. demonstrated how scRNA-seq in atherosclerotic lesions identified distinct endothelial subsets specializing in lipid handling, oxidative stress response, and leukocyte recruitment.1 Bondareva and Sheikh further highlighted that scRNA-seq platforms uncover vascular zonation patterns and region-specific endothelial responses to inflammation, thereby redefining our understanding of vascular homeostasis.2 In human heart failure, Rao et al. mapped fibrotic and non-fibrotic myocardial tissues, revealing that fibrotic-region ECs could upregulate adhesion molecules and foster leukocyte infiltration.3 Notably, tools for single-cell trajectory inference allowed dynamic modeling of endothelial activation, illustrating a continuum from quiescence to inflammation rather than discrete states.
In oncology, scRNA-seq has improved the concept of tumour-associated ECs (TECs). Shiau et al. performed single-nucleus RNA-seq on pancreatic ductal adenocarcinoma samples and identified a “reactive EndMT” program enriched in hypoxia-driven signaling and mesenchymal traits, which correlates with poor outcomes.4 Yang et al. integrated scRNA-seq-derived TEC markers into a liver cancer prognostic model and then linked endothelial gene signatures to immune infiltration and therapy response.5 In gastric cancer, Chen et al. used longitudinal scRNA-seq to reveal endothelial expansion and pro-angiogenic activation following neoadjuvant chemotherapy,6 suggesting that endothelial remodeling is highly dynamic and therapy-responsive.
Recent studies further highlight the critical role of vascular ECs in cancer progression. Using single-cell RNA sequencing, Zhang et al. characterized endothelial heterogeneity in colorectal cancer liver metastases and identified specific subpopulations that actively engage with immune cells.7 In gastric cancer, Yang et al. demonstrated that chemotherapy dynamically alters endothelial-immune crosstalk within the tumour microenvironment.8 In sum, these findings establish ECs as key regulators of both vascular structure and immune modulation in tumours.
Beyond disease states, single-cell atlases have deepened our understanding of tissue-specific endothelial specialization. In human skin, He et al. identified six distinct blood vascular endothelial subtypes, each demonstrating specialized patterns of metabolism and immune modulation.9 Li et al. extended this outcome by showing that capillary ECs in the skin exhibited strong HLA-II expression profiles, suggesting active participation in immune surveillance.10 These dermal vascular maps reinforce the concept that endothelial identity is shaped by environmental cues and localized tissue demands.
At the systems level, Augustin and Koh suggested rethinking the vascular endothelium as a widely distributed organ that actively shapes organ function, rather than merely acting as a passive barrier.11 This new perspective is increasingly supported by single-cell studies showing that ECs can secrete regulatory signals, adjust their metabolism, and remodel their local tissue environment in response to stress.
Looking ahead, vascular research will increasingly depend on integrated single-cell multi-omics approaches—including transcriptomics, chromatin accessibility, proteomics, and spatial mapping—to fully capture the complexity of endothelial biology in both health and disease. As summarized by Shiau et al. and further expanded by Bondareva and Sheikh, ECs are inherently plastic, swiftly adapting to mechanical stress, metabolic changes, inflammatory cues, and therapeutic interventions.2, 4 The predictive power of scRNA-seq, especially when combined with trajectory analysis and intercellular communication analysis, firmly positions it as a cornerstone for precision vascular medicine.
Beyond cataloging static cell types, single-cell technologies are evolving towards dynamic profiling of endothelial transitions over time. Longitudinal single-cell analyses have the potential to track how ECs shift during disease initiation, progression, and therapeutic response, moving our understanding from isolated snapshots to continuous biological narratives. Integrating complementary technologies, such as single-cell ATAC-seq to probe chromatin landscapes, spatial transcriptomics to preserve tissue architecture, and single-cell proteomics for functional validation, will further enhance mechanistic insights into endothelial regulation.
To fully realize this potential, the development of comprehensive single-cell atlases across diverse populations, including pediatric, aging, and ethnically varied cohorts, will be critical for uncovering context-dependent vascular adaptations. Coupling large-scale single-cell datasets with machine learning may enable the prediction of endothelial states, disease trajectories, and therapeutic vulnerabilities. Ultimately, single-cell technologies not only promise to redefine vascular biology with unprecedented resolution but also to pave the way for precision interventions targeting endothelial dysfunction across a broad range of diseases.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.




