Current advances in the role of classical non-homologous end joining in hematologic malignancies
Abstract
Background
Double-strand breaks (DSBs) are universally acknowledged as the most detrimental type of DNA damage, and their effective repair primarily depends on the non-homologous end joining (NHEJ) pathway. Such DSBs, which require NHEJ for resolution, can arise from intrinsic and extrinsic DNA-damaging factors or emerge naturally during essential biological processes like V(D)J recombination and antibody class switch recombination.
Main Body
Failure to properly repair DSBs may lead to genomic instability, disruption of cellular functions, and immunodeficiency, thereby promoting the development of hematologic malignancies. Conversely, overexpression of NHEJ-related genes can enhance resistance to DNA-damaging therapies in these cancers. Analyzing mutations in key classical NHEJ (cNHEJ) components and understanding their mechanisms could provide valuable biomarkers for predicting therapeutic outcomes and guiding treatment decisions. Consequently, defects in cNHEJ may offer insights into the development of novel drugs targeting DNA repair pathways.
Conclusion
We focus on genetic changes and alterations in gene regulation, while also providing an overview of cNHEJ.
1 BACKGROUND
Among all forms of DNA damage, double-strand breaks (DSBs) pose the greatest threat due to their potential to cause extensive chromosomal loss, with a substantial number of these lesions being resolved through the non-homologous end joining (NHEJ) mechanism.1 In mammalian cells, classical NHEJ operates as a swift and high-capacity repair system that ligates DNA ends with minimal sequence alignment, often resulting in mutations at the repair sites.2 This pathway is indispensable for repairing both pathological DSBs and physiological breaks occurring during V(D)J recombination and immunoglobulin heavy chain class switching.1-3 Furthermore, defective NHEJ can trigger premature apoptosis in lymphocytes, highlighting its critical role in preserving genomic integrity and supporting immune system functionality.2 (See Figure 1)
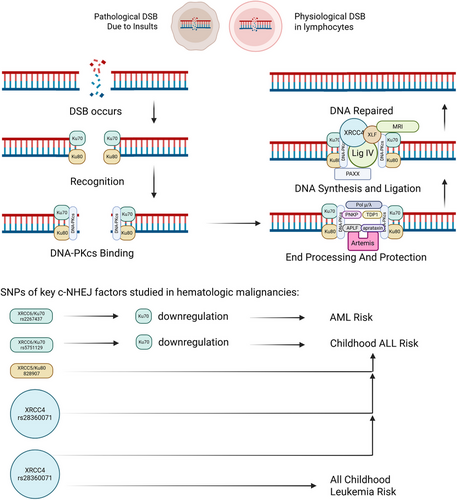
Hematologic malignancies, which include leukaemia, lymphoma, and multiple myeloma (MM), constitute a heterogeneous group of cancers with distinct biological and clinical characteristics. The advent of genomic profilling has significantly transformed the landscape of diagnosis, prognosis, and treatment strategies for these diseases, particularly through the identification and analysis of DNA repair pathways.4 These genetic mechanisms, which play a crucial role in maintaining genomic integrity, hold immense potential as diagnostic and prognostic indicators. Further research into these pathways is essential to refine their application in disease classification, risk stratification and the development of targeted therapies. By leveraging these insights, clinicians can enhance precision medicine approaches, optimising therapeutic outcomes for patients with hematologic malignancies.
This review delves into the role of classical non-homologous end joining (cNHEJ) in the initiation and advancement of hematologic malignancies. Additionally, it emphasises the impact of genetic changes in core cNHEJ components on the formation and clinical outcomes of leukaemias, lymphomas, and myelomas.
1.1 Classical non-homologous end joining general introduction
The Ku70/80 complex serves as the primary sensor for DSBs, binding to the damaged DNA ends and initiating the recruitment of downstream NHEJ components. DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is subsequently recruited, displacing Ku and positioning itself at the DNA termini to facilitate further repair. Key scaffolding proteins, including X-ray repair cross complementing protein (XRCC4), DNA ligase IV (LigIV) and XRCC4-like factor (XLF), assemble at the site, enabling end-bridging through intricate protein–protein interactions, while Artemis processes hairpin DNA ends to prepare them for repair. Opposing DNA-PKcs molecules undergo trans-autophosphorylation, leading to structural reorganisation of the complex and eventual dissociation of DNA-PKcs, thereby exposing the DNA ends for processing and ligation. PAXX interacts with XRCC4 and XLF, providing additional structural stability to the complex and functioning redundantly with XLF to reinforce the scaffolding network. PNKP, a human kinase and phosphatase, and TDP1, an enzyme, play a role in processing tricky DNA ends, paving the way for ligation. Besides, polymerases like Polμ, Polλ interacts with the complex by fitting into gaps in non-complementary ends, so as to lower the extent of deletions. Artemis is considered to be the major nuclease in cNHEJ, as other nucleases, namely aprataxin and APLF, also play their roles in the reparation.1-3, 5, 6 Furthermore, MRI/Cyren, a cell-cycle-specific inhibitor of cNHEJ, plays a dual role by enhancing the retention of cNHEJ factors and compensating for the absence of XLF in lymphocytes, ensuring repair efficiency.7, 8
1.2 Ku70/80 heterodimer
Ku70 and Ku80 are synthesised from the XRCC6 and XRCC5 genes. During the initial step of DNA end recognition, the Ku complex detects DSBs, binds tightly to the DNA termini, protects them from further damage, and signals other NHEJ proteins to join the repair process.6, 9
Ku70, widely studied for its role in DNA repair, also participates in diverse biological processes such as aging, cellular senescence, apoptosis, and the replication of certain viruses. Additionally, research has revealed its function in innate immunity, where it acts as a pattern recognition receptor. Upon detecting viral DNA in human cells, Ku70 initiates an antiviral immune response by inducing the release of interferons and pro-inflammatory cytokines.10
Studies on knockout mice demonstrate that deleting either the Ku70 or Ku80 gene in p53-mutated mice results in similar phenotypic consequences, including increased sensitivity to clastogens, accelerated aging, and a heightened risk of pro-B-cell lymphoma, although the impact of Ku70 deletion is less pronounced compared to Ku80.11 Earlier investigations in mice further indicate that the deletion of one or both genes does not significantly elevate lymphoma incidence but is strongly associated with premature aging.12 Additionally, Ku80/p53 double-deficient mice exhibit reduced resistance to experimentally induced medulloblastoma, while DAOY cells, a human medulloblastoma cell line, are naturally devoid of nuclear Ku70.13, 14 Despite these findings in murine models, there remains a lack of robust and conclusive evidence regarding the effects of Ku70 or Ku80 gene knockout in human cells, highlighting the need for further research in this area.
Through targeted gene sequencing, a study identified altered expression of XRCC5 and XRCC6 in B-cell lymphoma samples, particularly diffuse large B-cell lymphomas (DLBCLs). These changes led to defects in NHEJ, contributing to genomic instability in these tumours.15 Similar findings were observed in human pre-B cell samples associated with leukaemia, where heterozygous Ku70 mutant pre-B leukaemic cells exhibited increased sensitivity to DNA-damaging agents, a preference for microhomology-mediated end joining (MMEJ), and elevated markers of cellular aging following radiation exposure.16
Recent research has highlighted Ku70 as a critical factor linked to the frequency of chromosomal translocations in human lymphocytes after radiation exposure and in T-cell acute lymphocytic leukaemia (ALL). This connection suggests that Ku70 could serve as a valuable biomarker for assessing radiation-induced damage and may also emerge as a potential therapeutic target in the cancer treatment.17
Additionally, a cross-sectional study involving 95 acute myeloid leukaemia (AML) patients revealed that reduced expression of XRCC6, the gene encoding Ku70, correlated with improved prognosis.18
Single nucleotide polymorphisms (SNPs) are naturally occurring variations in the DNA sequence that are prevalent within populations and can significantly impact gene regulation and functional outcomes. This study further identified that SNP rs2267437, in the XRCC6 gene can suppress its expression, leading to reduced levels of its encoded protein, Ku70, as well as a worsened prognosis in AML.18 Besides, in acute lymphoblastic leukaemia, SNP rs5751129 of XRCC6 suppresses its trasnscription, resulting in an augmented odds; for XRCC5 rs828907, a higher odds was observed too.19 Another study highlighted that acetylation of Ku70 can inhibit the NHEJ repair pathway, thereby promoting apoptosis. Emerging evidence suggests that Ku70 is a potential target for histone deacetylase inhibitors and plays a pivotal role in the induction of programmed cell death.20 Additionally, OTUD5, a specific deubiquitinase for Ku80, enhances its stability and functions as a positive regulator of NHEJ repair, ensuring efficient DNA damage response.21 In a separate investigation, it was discovered that TOX interferes with the binding of the Ku70/80 complex to DNA damage sites, impairing NHEJ and promoting genomic instability in T-cell ALL.22
1.3 DNA-dependent protein kinase catalytic subunit
The DNA-PK complex, a heterotetramer with a molecular weight of around 650 kDa, is formed by the association of DNA-PKcs, Ku70/80, and a DNA duplex. DNA-PKcs, encoded by the PRKDC gene and a member of the phosphatidylinositol 3-kinase-related kinase family, is the second factor recruited in the NHEJ pathway. This complex is essential for activating and phosphorylating a wide array of proteins involved in the repair process.5, 6
Studies in mice reveal that DNA-PKcs-null mutations result in severe combined immunodeficiency (SCID), marked by the absence of mature B and T cells due to impaired V(D)J recombination.23 The absence of DNA-PKcs also promotes MMEJ and increases chromosomal translocations, which drive the development of cancers such as leukaemias and lymphomas.24 Mice lacking both PARP1 and DNA-PK exhibit a higher incidence of T-cell lymphoma, driven by p53 mutations and telomere fusion.25, 26 Additionally, a human case with a DNA-PKcs missense mutation showed defective Artemis activation and impaired NHEJ.27
Recent studies have delved deeper into the functional roles of kinases, particularly their catalytically inactive forms. For instance, DNA-PKcs, when rendered inactive, has been shown to induce higher levels of genomic instability compared to its complete absence. This observation highlights the intricate challenges and potential risks associated with developing and deploying targeted kinase inhibitors, such as those aimed at DNA-PKcs, in the cancer treatment. It underscores the critical importance of precision in therapeutic approaches to avoid unintended consequences.28
Complementing this perspective, a bioinformatics analysis revealed that DNA-PK activation is prevalent in the majority of AML blast samples, positioning it as a viable therapeutic target in AML. The study suggests that inhibiting DNA-PK could disrupt cancer cell survival mechanisms and enhance the effectiveness of existing treatments. In clinical practice, combining multikinase inhibitors with DNA-PK-specific inhibitors may provide substantial therapeutic advantages, potentially improving patient outcomes and addressing resistance to conventional therapies.29
In the context of melanomas, recent research indicates that suppressing the DNA-PK activity can enhance tumour immunogenicity by upregulating neoantigen expression and presentation while expanding the population of neoantigen-reactive T cells. However, whether this effect translates to hematologic cancers remains to be validated.30
Additionally, DNA-PK has been implicated as a transcriptional regulator, driving the expression of target enzymes that contribute to anthracycline chemoresistance in AML. This finding further emphasises the multifaceted role of DNA-PK in cancer biology and its potential as a therapeutic target.31
DNA-PK inhibitors have emerged as promising therapeutic agents in hematologic malignancies, with distinct effects across different leukaemia subtypes. AZD-7648, a potent DNA-PK inhibitor, has been shown to exert multiple anti-tumour effects in vitro, including the inhibition of cell multiplation, induction of DNA lesion, activation of apoptotic pathways, and stasis of the cell cycle in AML and chronic myeloid leukaemia (CML) cells. However, its therapeutic impact appears to be less pronounced in ALL, as evidenced by findings from a separate study.32, 33 Meanwhile, CC-115, a dual inhibitor targeting both TORK and DNA-PK, has shown clinical potential in chronic lymphocytic leukaemia (CLL) by disrupting critical signalling pathways essential for cell proliferation.34 Peposertib (M3814), another DNA-PK inhibitor, effectively blocks the NHEJ pathway, enhancing p53-mediated cytotoxicity in acute leukaemia when combined with therapies that induce DSBs, such as ionising radiation or topoisomerase II inhibitors.35 Additionally, in pre-B ALL cell experiments, the combination of the DNA-PK inhibitor NU7441 and the telomerase inhibitor MST-312 significantly increased apoptosis rates, suggesting a potential therapeutic strategy for this subtype.36
In MM, inhibition of DNA-PKcs could enhance the efficacy of a DNA-damaging-inducing agent doxorubicin, but caused by immunity-related pathway rather than defective cNHEJ.37
In DLBCL, somatic mutations in DNA-PKcs have been observed.38
1.4 Artemis
The activation of Artemis, a 5′ to 3′ endonuclease crucial for resolving hairpin DNA structures, is mediated by DNA-PKcs.5, 31 Together, these proteins are integral to V(D)J recombination, a process vital for the development of lymphoid cells and the establishment of a functional immune system.38-41
In addition, Artemis has been identified as a tumour suppressor, with its deficiency associated with immunodeficiency and an increased risk of developing lymphoma and non-lymphoid malignancies.42 In the context of ALL, targeting Artemis activity has emerged as a potential therapeutic strategy, as its inhibition can effectively suppress the proliferation of B- or T-ALL cells. However, this approach may come at the cost of compromised immune function due to its broader role in the lymphocyte development.33 Experimental studies have demonstrated that pharmacological inhibition of Artemis significantly reduces the growth of B-ALL cell lines, while mature B-cell lines remain largely unaffected, highlighting its selective impact on immature lymphoid cells.33 Structural investigations into the molecular architecture of Artemis have provided valuable insights, paving the way for the design of specific inhibitors that could enhance therapeutic precision in ALL treatment.43 (See Table 1
| Inhibitors | Target | Disease | Therapy | Study type | Reference |
|---|---|---|---|---|---|
| AZD-7648 | DNA-PK | CML/AML | Alone | Cell test | 32, 33 |
| CC-115 | DNA-PK/TORK | CLL | Alone | Clinical trial | 34 |
| Peposertib (M3814) | DNA-PK | Acute Leukaemia | With ionising radiation or topoisomerase II inhibitors | Clinical trial | 35 |
| NU7441 | DNA-PK | Pre-B ALL | With telomerase inhibitor MST-312 | Cell test | 36 |
| Compound 827171 | Artemis | ALL | Alone | Cell test | 33 |
| Compound 827032 | Artemis | ALL | Alone | Cell test | 33 |
| Compound 826941 | Artemis | ALL | Alone | Cell test | 33 |
| Compound 825226 | Artemis | ALL | Alone | Cell test | 33 |
In DLBCL, the expression of Artemis has been observed to be altered selectively.15
1.5 LigIV–XRCC4–XLF complex
The final step is implemented by the LigIV–XRCC4–XLF complex, which ligates the DNA ends.5
1.6 DNA ligase IV
Deficiency in LigIV has been shown to trigger extensive neuronal apoptosis and halt lymphocyte development, ultimately leading to late embryonic lethality in mice through the p53 pathway. However, mice with mutations in both LigIV and p53 are viable, exhibiting impaired lymphogenesis, a predisposition to pro-B-cell lymphomas, and an increased risk of medulloblastoma.44-47
In a clinical case, a patient with LigIV deficiency presented with SCID. The absolute count of B cells was drastically reduced by 100-fold, while alphabeta T cells decreased by 10-fold, primarily due to defects in mechanisms responsible for generating immune diversity. These defects contribute to an elevated risk of tumourigenesis, particularly lymphoma and leukaemia.48 Furthermore, recent evidence suggests that monoallelic LigIV missense mutations, through haploinsufficiency, can lead to immune dysregulation in humans, manifesting as autoimmunity or immunodeficiency.49
Though LigIV SNP rs1805388 suppresses the expression of itself, no significant association with prognosis was observed in AML.18
1.7 X-ray repair cross complementing protein 4
Similar to LigIV deficiency, the absence of XRCC4 in mice leads to severe consequences, including embryonic lethality, widespread neuronal apoptosis, and impaired cellular proliferation. However, mice with double deficiencies in both XRCC4 and p53 can circumvent these effects, though they exhibit defective V(D)J recombination and impaired the lymphocyte development. These mice are predisposed to developing pro-B-cell lymphomas, likely due to heightened genomic instability.50 Additionally, the combined deficiency of XRCC4 and p53 accelerates the onset of medulloblastoma, further underscoring the role of XRCC4 in maintaining genomic stability.51
XRCC4 has also been implicated in modulating apoptosis induced by chemotherapy agents such as paclitaxel and vincristine. This suggests that the regulation of XRCC4 could serve as a predictive biomarker for evaluating the efficacy of apoptosis-inducing therapies in the cancer treatment.52
Findings indicate that genomic variants of XRCC4 genotypes may influence susceptibility to CML, and MM.53, 54 Global studies have explored the association between SNPs in XRCC4 and hematologic cancer risk. In childhood ALL, XRCC4 rs6869366 or XRCC4 rs28360071 are tied with increased odds, though the mRNA levels of XRCC4 are not altered; while in all childhood leukaemia, rs6869366 is also considered as a risk factor.19, 55
1.8 XRCC4-like factor
Unlike other NHEJ factors, XLF deficiency in mice leads to a gradual decline in lymphocyte levels, which is generally less severe than SCID. Interestingly, XLF-deficient lymphocytes in mice do not exhibit significant defects in V(D)J recombination.56 This phenomenon may be attributed to functional redundancy between XLF and other NHEJ components or DNA damage repair factors, such as PAXX and 53BP1.1, 56-59 However, mature B cells lacking XLF display moderate impairments in immunoglobulin heavy-chain class switch recombination. Mice deficient in both XLF and p53 do not show a pronounced predisposition to pro-B-cell lymphomas but are prone to developing medulloblastomas.60
In humans, XLF deficiency can cause microcephaly, delayed neural development, and varying degrees of immune deficiency, characterised by reduced lymphocyte counts and increased radiosensitivity. Despite sharing the same genetic defect, clinical phenotypes and immunological profiles can vary significantly among individuals.61-63
XLF- or PAXX-deficient mice are viable and exhibit only mild immune system impairments. However, mice lacking both genes experience embryonic lethality due to neuronal apoptosis, genomic instability, and nearly complete arrest of lymphogenesis. Studies have shown that XLF and PAXX collaborate in V(D)J recombination, while PAXX plays a critical and independent role in class switch recombination.64-67 Remarkably, triple-deficient mice lacking XLF, PAXX, and p53 are viable but display a dramatic reduction in mature lymphocyte populations and a significant increase in pro-B-cell numbers.68, 69 Additionally, combined deficiency of XLF and MRI results in embryonic lethality,69, 8 whereas selective depletion of MRI in mice reduces class switch recombination activity in B lymphocytes and slows the proliferation of neuronal progenitor cells.70
2 CONCLUSION
In summary, this review synthesises recent advancements in understanding the role of the cNHEJ pathway in hematologic malignancies, with a particular emphasis on its core molecular components and their functional implications. cNHEJ is a critical DNA repair mechanism that maintains genomic stability by repairing DSBs. However, this process is not error-free and can introduce mutations, which may contribute to oncogenesis. Beyond its role in DNA repair, NHEJ is indispensable for V(D)J recombination, a fundamental process in the lymphocyte development that generates the diversity of antigen receptors essential for adaptive immunity. The precise regulation of NHEJ genes is therefore vital for both responding to internal and external DNA damage and ensuring the proper maturation of lymphocytes.
In hematologic malignancies, NHEJ factors are often selectively mutated or dysregulated, and specific polymorphisms in these genes can influence an individual's susceptibility to such cancers as well as their clinical outcomes. Severe deficiencies or complete loss of certain NHEJ genes, such as LigIV or XRCC4, frequently result in genomic instability and profound immune deficiencies, often manifesting as lymphomas or leukaemias. Notably, the absence of these genes can lead to embryonic lethality in mice, primarily through activation of the p53 pathway. Additionally, functional redundancy exists among some NHEJ factors, as demonstrated by XLF and PAXX. While mice deficient in either XLF or PAXX alone are viable, simultaneous deficiency in both genes results in embryonic death due to massive neuronal apoptosis, genomic instability, and arrested lymphogenesis.
In the context of hematologic tumours, elevated expression of NHEJ genes may enhance genomic stability, thereby conferring resistance to therapies that rely on DNA-damaging agents, such as chemotherapy or radiation. This observation suggests that the expression levels of specific NHEJ genes could serve as valuable biomarkers for predicting prognosis and tailoring treatment strategies. For example, combining DNA-damaging drugs with inhibitors targeting specific NHEJ components may enhance therapeutic efficacy by increasing tumour cell vulnerability. Several such inhibitors are currently under development, with some advancing to clinical trials. However, further research and clinical validation are needed to fully understand their potential and limitations.
Moreover, the functional interplay between NHEJ factors and other DNA repair pathways, such as MMEJ, adds another layer of complexity to their roles in cancer biology. Investigating these interactions could uncover novel therapeutic targets and strategies for overcoming treatment resistance in hematologic malignancies. Additionally, the development of biomarkers based on the NHEJ gene expression or mutational status could facilitate personalised treatment approaches, enabling clinicians to select the most effective therapies for individual patients.
In conclusion, the cNHEJ pathway plays a multifaceted role in hematologic malignancies, influencing genomic stability, immune function and treatment resistance. While significant progress has been made in understanding its mechanisms and therapeutic potential, further research is needed to translate these insights into clinical practice. By leveraging the unique characteristics of NHEJ factors, researchers and clinicians may be able to develop more precise and effective treatments for hematologic cancers, ultimately improving patient outcomes.
AUTHOR CONTRIBUTIONS
Conceptualisation: Zizhen Xu and Pengcheng Liu. Data curation: Pengcheng Liu and Zizhen Xu. Formal Analysis: Pengcheng Liu. Funding Acquisition: N/A. Investigation: Pengcheng Liu and Zizhen Xu. Methodology: Pengcheng Liu and Zizhen Xu. Project administration: Zizhen Xu. Resources: Zizhen Xu. Software: Pengcheng Liu and Zizhen Xu. Supervision: Zizhen Xu. Visualisation: Pengcheng Liu. Original Draft: Pengcheng Liu. Review & Editing: Zizhen Xu and Pengcheng Liu.
ACKNOWLEDGMENTS
The authors have nothing to report.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
This work is funded by National Natural Science Foundation of China No. 81600151.
ETHICAL APPROVAL
None.




