Ubiquitin-dependent regulation of MYC: A promising therapeutic target
Commentary on:
Xiong Y, Wang L, Xu S, Fu B, Che Y, Zaky MY, Tian R, Yao R, Guo D, Sha Z, Lin F, Lin X, Wu H. Small molecule Z363 co-regulates TAF10 and MYC via the E3 ligase TRIP12 to suppress tumour growth. Clin Transl Med. 2023;13(1):e1153. doi: 10.1002/ctm2.1153
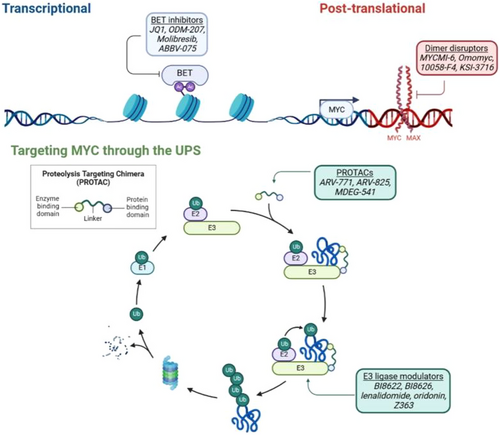
MYC is a pleiotropic transcription factor involved in the regulation of critical cellular processes including cell proliferation, differentiation and apoptosis. Expression and activity of MYC are deregulated in over 70% of human malignancies, contributing to both tumour initiation and maintenance, and are frequently associated with poor prognosis. This central role in oncogenesis makes MYC a highly desirable therapeutic target. Direct targeting of MYC on a protein level is challenging due to the disordered nature of the protein structure and lack of enzymatic activity, nonetheless, the first direct inhibitor Omomyc, a dominant-negative mutant of MYC, recently entered clinical trials.1 In addition, numerous alternative strategies to target MYC transcription, translation, stability and interaction with dimerization partner MAX are also under investigation. One promising approach to indirectly target MYC is to pursue molecules responsible for its post-translational regulation (Figure 1). To date, at least 18 E3 ligases have been identified to mediate MYC ubiquitination, functioning to either regulate MYC stability or activity and are considered potentially exploitable targets. For example, the natural diterpenoid compound oridonin activates the E3 ligase SCFFBW7 leading to targeted degradation of MYC through the proteasome.2 On the other hand, inhibition of the E3 ligase HUWE1, which acts to promote MYC function, results in suppression of MYC-dependent transactivation, blocks the formation of protective MYC multimers at the replication fork and enhances MYC degradation.3-5 Another emerging strategy to promote MYC degradation is using proteolysis targeting chimaeras (PROTACs). These are heterobifunctional small molecules that simultaneously bind a target and an E3 ligase to induce the ubiquitination and degradation of the target protein. So far, no PROTACS have been developed to induce direct degradation of MYC, but PROTACS targeting bromodomain and extra-terminal domain (BET) proteins indirectly reduce MYC expression and show promising pre-clinical activity.6, 7
A recent study from Xiong et al. identified a small molecule, Z363, that promotes the degradation of MYC and TAF10 via the E3 ligase TRIP12. The authors demonstrated elevated expression of MYC and the transcription factor TAF10 across a range of primary tumour types when compared to adjacent non-tumour tissues and hypothesised that TAF10 could enhance the transcriptional activity of MYC. Accordingly, knockdown and overexpression studies led to decreased MYC expression and increased MYC promoter activity, respectively. Mutation studies in the breast cancer cell line MCF7 identified that the MBIV domain on MYC was essential for this interaction. The MBIV domain has been reported to enhance the transcription of 150 genes, however, the effects of deletion of this domain were variable across model systems suggesting that it may only be required for transformation in specific biological contexts.8 Further understanding of the functional significance of this domain across breast cancer and other cancer types would be of great interest.
Xiong and colleagues next screened a panel of novel small molecules against MCF7 cells and identified that compound Z363 led to decreased expression of both MYC and TAF10. The proteasome inhibitor MG132 rescued the expression of both proteins indicating that Z363 promotes their degradation through the proteasome. The authors employed an online tool (UbiBrowser) to predict potential E3 ligases that can target MYC and TAF10. From this, 11 candidate E3 ligases were identified and subsequent knockdown studies demonstrated that TRIP12 could modulate protein levels of both MYC and TAF10. TRIP12 expression increased in response to treatment with Z636 and both Z363 and TRIP12 overexpression similarly inhibited proliferation and promoted apoptosis in MCF7 cells. TRIP12 decreased MYC stability by first modulating MYC phosphorylation at T58 to promote subsequent degradation by TRIP12. Comparatively, oridonin targets MYC through upregulation of the kinase GSK-3 and E3 ligase SCFFBW7 to induce increased T58 phosphorylation and MYC degradation, respectively.3 It will be important for future studies to delineate the intermediates involved in promoting T58 phosphorylation in response to Z363 and investigate whether there is functional redundancy between TRIP12- and SCFFBW7-catalysed ubiquitination of MYC. Z363 was also found to promote TRIP12-mediated degradation of the transcription factor TAF10. This draws similarities with the mode of action of immunomodulatory drugs (IMiDs) which promote CRL4CRBN-mediated ubiquitination of transcription factors IKZF1 and IKZF3 resulting in downregulation of MYC activity and further highlights the therapeutic promise of Z363. Together the findings from this study indicate a model whereby TRIP12 and TAF10 dynamically regulate MYC transcription and protein stability and present Z363 as a potential novel anti-cancer strategy which co-regulates both proteins to suppress MYC.
TRIP12 is a HECT domain E3 ligase that is best known for its role in the cell cycle through regulation of the tumour suppressor p14/ARF. It is also implicated in the regulation of cellular processes such as chromatin remodelling, DNA repair and cell differentiation. TRIP12 expression is reported to be overexpressed in a number of cancers, including breast cancer, and has been proposed as a potential therapeutic target.9 Xiong and colleagues introduce TRIP12 as a novel player in regulating both MYC stability and transcriptional activity and identify the small molecule Z363 as an activator of TRIP12 activity. This work encourages further investigation to delineate both the role of TRIP12 in tumourigenesis and the mode of action of Z363. One key avenue for exploration will be to understand the relative contributions of TRIP12 and other E3 ligases in regulating MYC stability and whether they act in a cell type-specific manner. Furthermore, it will be imperative to investigate whether Z363 affects TRIP12-mediated ubiquitination of other known substrates, particularly p14/ARF. Intriguingly, TRIP12 was recently identified as a key promoter of PROTAC-mediated degradation of the BET protein BRD4. TRIP12 functions co-operatively with CRL complexes to generate K29/K48-branched ubiquitin chains, accelerating the degradation of BRD4 which in turn leads to suppression of MYC activity.10 It would be fascinating to explore whether PROTAC-based therapies exhibit enhanced efficacy in tumours with high expression of TRIP12 or indeed whether activation of TRIP12 by Z363 can increase the efficacy of PROTACs. In summary, targeting post-translational regulation of MYC is a promising anti-cancer strategy, however, given the myriad of different pathways involved in regulating MYC activity and stability, a better understanding of this dynamic control and crosstalk will be important to progressing therapeutic strategies.




