Recent development of urinary biomarkers for bladder cancer diagnosis and monitoring
Yuchen Zeng, Anqi Wang, Wei Lv and Qingqing Wang contributed equally.
Abstract
Urine-based liquid biopsy has emerged as a non-invasive and effective tool for early screening and diagnosis of bladder cancer. This review provides a comprehensive overview of the current urine-based biomarkers and methods for the detection and monitoring of bladder cancer. We focus on biomarkers including tumour DNAs, proteins, microbiome, tumour RNAs, long non-coding RNAs, transfer RNA-derived fragments, messenger RNAs, microRNAs, circular RNAs, exosomes and extrachromosomal circular DNA.
1 INTRODUCTION
Bladder cancer (BC) mainly occurs on the mucous membrane of the bladder and is one of the most lethal urologic malignancies.1 Based on histological classifications, BC can be classified into urothelial bladder carcinoma (UBC), squamous cell carcinoma and adenocarcinoma. Notably, more than 90% of BCs are UBCs.2 Moreover, according to the depth of tumour invasion, BC is clinically categorised into non-muscle invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC).2 About 75% of BCs are NMIBCs,3 which have an estimated 5-year survival rate of 77% and an extremely high recurrence rate of 50%.4 When BC progresses into the metastatic stage, its overall 5-year survival rate reduces to approximately 5%. Thus, early detection of BC is urgently needed to improve patients' survival and quality of life.
Symptoms of haematuria, frequent urination and recurrent urinary tract infections are the hallmarks of BC.5 Screening methods for BC include both invasive and non-invasive procedures. Clinicians usually choose a combination of non-invasive urine biopsies and imaging tools for early screening of BC.5 Imaging tests including ultrasonography, computed tomography and magnetic resonance imaging are regularly chosen for further examinations. Cytology and cystoscopy are the current gold standard methods for BC screening.5 Cystoscopy has high BC-detection specificity; however, it is a highly invasive and low patient compliance method.6 Urine cytology, which was widely used to detect and monitor BC, is a non-invasive test with a high specificity of 85%–100%, but suffers from intrinsic limitations, such as low dynamic ranges and low inherent sensitivity (13%–75%).5 Thus, urine cytology has a limited clinical application value in the detection of low-grade BC.7 In addition, the diagnostic accuracy of the urine cytology test is also subjective and mainly depends on the expertise of the examining physician.8 The outcome of urine cytology can be influenced by inflammation and infection of the urinary system, which leads to false-positive results.
Urine is an ideal clinical source for fluid biopsy and has the advantages of being cost-effective and non-invasive.9 Urine-based liquid biopsy is routinely used in urine examinations, urine cytology and urinary tumour biomarkers.10 Urine-based liquid biopsy has notable advantages over cystoscopy in avoiding side effects. Also, it is suitable for high-frequency sampling during early screening and BC prognosis. However, improvements are still needed to increase the accuracy of urine-based liquid biopsy for the early screening and diagnosis of BC. This review summarises current progress in developing urinary tumour biomarkers for BC diagnosis and monitoring, including urinary tumour DNA/RNA, proteins, microbiome, long non-coding RNAs (lncRNAs), transfer RNA-derived fragments (tRFs), messenger RNAs (mRNAs), microRNAs (miRNAs), exosomes, circular RNAs (circRNAs) and extrachromosomal circular DNA (eccDNA). We also highlight the advantages and limitations of urine-liquid biopsy in the diagnosis of BC and the techniques related to urine-based liquid biopsy (Figure 1).
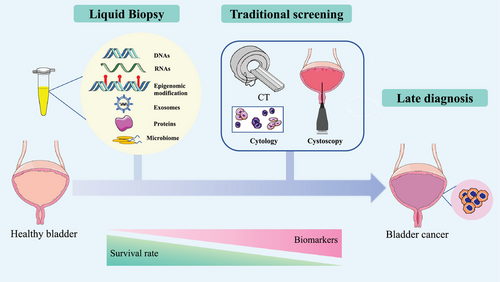
2 URINE-BASED LIQUID BIOPSY
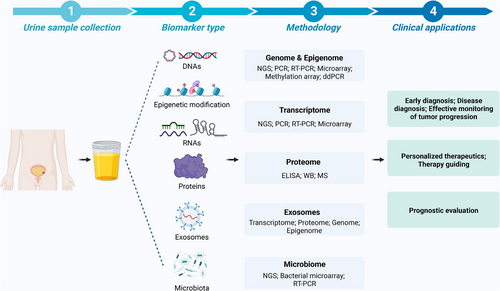
Liquid biopsy mainly refers to blood or urine specimens and it has emerged as a powerful non-invasive method for cancer diagnosis and monitoring.11 In the past decades, liquid biopsy has been widely applied in multiple cancer types.12, 13 It can avoid the complexity of tissue biopsy and acquire genomic information of tumour at an early stage by using a simple method. Tumour cells could release large amounts of tumour-associated biomolecules into the circulation system, and part of them could be further filtered into the urine through the glomerular filtration barrier. Moreover, the special physiological structure of BC allows the direct contact of tumours with urine. Therefore, urine-based liquid biopsy is regarded as a reliable and convenient source for the detection of urogenital disorders (Figure 2 and Table 1).10
| Biomarkers | Cohorts | Methods | Efficiency | Clinical application | Ref. |
|---|---|---|---|---|---|
| DNAs | |||||
| ucfDNA | 103 NMIBC patients | RT-PCR | NA | Prognosis and recurrence prediction for NMIBC | Xu et al.28 |
| uctDNA | 12 NMIBC patients | RT-PCR | SN = 83% | Diagnose and monitor NMIBC | Birkenkamp-Demtröder et al.35 |
| ucfDNA | 42 BC patients, 27 healthy controls | NGS, uCAPP-seq | SN = 81%, SP = 81% | Early detection of MIBC | Chauhan et al.47 |
| ucfDNA | Test cohort: 74 pre-TURBT patients and 52 haematuria group; validation cohort: 40 pre-TURBT patients and 36 surveillance group | ddPCR, RT-PCR | Test cohort: SN = 68.9%, SP = 96.2%; validation cohort: SN = 77.5% | Diagnosis of UBC | Hayashi et al.42 |
| ucfDNA | 126 UC patients, 64 healthy controls | UroCAD | SN = 82.5%, SP = 96.9% | Diagnosis of UC | Zeng et al.50 |
| ucfDNA | 66 BC patients, 34 controls | RT-PCR | Low grade: SN = 20.7%, SP = 91.2%; high grade: SN = 59.5%, SP = 91.2% | Diagnosis of BC | Brisuda et al.51 |
| ucfDNA | 12 BC patients, 10 healthy controls | NGS, RT-PCR, Sanger sequencing, 16S sequencing | SN = 91% | Diagnosis of BC | Ibrahim et al.55 |
| Urine microsatellite DNA | 103 BC patients, 80 other disease patients, 26 healthy controls | RT-PCR | SN = 80% | Diagnosis of BC in early stage | Schneider et al.57 |
| Epigenetic features | |||||
| Urine DNA methylation | 14 BC patients, 12 benign haematuria patients | Methylation-specific RT-PCR | SN = 78.6%, SP = 91.7% | Diagnosis of BC | Hentsche et al.23 |
| Urine DNA methylation | 44 BC patients, 83 healthy controls | Methylation-specific RT-PCR | Overall: SN = 85.4%, SP = 93.1%; T1: SN = 94.1%; T2: SN = 96.4%; T3: SN = 77.8%; T4: SN = 71.4% | Diagnosis of BC | Deng et al.60 |
| Urine DNA methylation | 72 BC patients, 75 healthy controls | Methylation-specific RT-PCR | SN = 92%, SP = 85% | Diagnosis of BC | Bosschieter et al.62 |
| Urine DNA methylation | 157 BC patients, 339 healthy controls | Methylation-specific RT-PCR | Two methylation markers panel: SN = 90%, SP = 93% | Diagnosis of BC | Renard et al.63 |
| Urine DNA methylation | 109 BC patients, 66 benign controls | utMEMA | Two methylation markers panel: overall SN = 90%, overall SP = 83.1%; minimal diagnosing SN = 81%; residual diagnosing SN = 93.3%; recurrent diagnosing SN = 89.5% | Diagnosis of low-grade BC, minimal BC, residual BC and recurrent BC | Chen et al.64 |
| Urine DNA methylation | 109 BC patients, 100 healthy controls | Methylation-specific RT-PCR | Two methylation markers panel: SN = 80%, SP = 93% | Diagnosis of high-stage BC | Hentschel et al.65 |
| Urine DNA methylation | 111 BC patients, 57 healthy controls | Pyrosequencing | Three methylation markers panel with cytology: SN = 96%, SP = 40% | Recurrence prediction of BC | van der Heijden et al.66 |
| Urine DNA methylation | 90 NMIBC patients | Pyrosequencing | Three methylation markers panel in testing group: SN = 86%, SP = 89% | Recurrence prediction of BC | Su et al.67 |
| Urine DNA methylation | 167 NMIBC patients, 105 healthy controls | Methylation-specific RT-PCR | Diagnosing: SN = 97.6%, SP = 84.8%; haematuria: SN = 90.3%, SP = 65.1% | Diagnosis and surveillance of NMIBC | Roperch et al.69 |
| Urine DNA methylation | 172 BC patients | Methylation-specific RT-PCR | Methylation marker with cytology: AUC = 0.773 | Diagnosis of BC | Fantony et al.68 |
| Urine cDNA histone modification | 300 AMH patients | RT-PCR | SN = 89%, SP = 63% | Diagnosis of BC | Lucca et al.72 |
| Proteins | |||||
| Urine protein | 101 BC patients, 109 benign urological disorders, 10 healthy controls | ELISA and Western blot | Urinary HIP/PAP levels: SN = 80.2%, SP = 78.2%; urinary NMP22 levels: SN = 52.1%, SP = 93.5%; urinary BTA levels: SN = 34.7%, SP = 96.1% | Early detection of BC, or recurrence prediction of NMIBC | Nitta et al.74 |
| Urine protein | 59 UBC patients, 57 healthy controls | ELISA, Western blot | NA | Diagnosis of BC | Allione et al.77 |
| Urine protein | 213 BC patients, 21 healthy controls | BC antigen, BTA stat test, NMP22 | BTA stat test: SN = 72.9%, SP = 64.6%; NMP22: SN = 63.5%, SP = 75.0%; BC antigen: SN = 80.5%, SP = 80.2% | Diagnosis of BC | Giannopoulos et al.80 |
| Urine protein | 111 NMIBC patients, 136 benign genitourinary disease, 50 healthy controls | Chemiluminescent immunoassay | Urine Ln-γ2m/uCRN AUC: 0.733 | Diagnosis of low-grade NMIBC | Karashima et al.83 |
| Urine cytokeratin | 109 Ta NMIBC patients | Immunohistochemistry, RT-PCR | NA | Early detection between LG and HG disease in Ta NMIBC | Muilwijk et al.85 |
| RNAs | |||||
| Urine mRNA | 61 BC patients, 37 controls | RT-PCR | ETS2:uPA RNA ratio: SN = 75.4%, SP = 100% | Early diagnosis of BC | Hanke et al.92 |
| Urinary cell-free RNA | 92 BC patients, 120 controls | RT-PCR | IQGAP3 levels: SN = 92.2%, SP = 65.2% (RiboGreen adjusted) | Diagnosis of MIBC and NMIBC | Kim et al.93 |
| Urinary cell-free RNA | 212 BC patients, 106 controls | RT-PCR | UBE2C levels: SN = 82.5%, SP = 76.2% | Diagnosis of BC | Kim et al.94 |
| Urine mRNA | 127 BC patients, 97 controls | RT-PCR, Western blot | NDRG2 levels: SN = 85.5%, SP = 81.4% | Diagnosis of BC | Zhang et al.95 |
| Urine mRNA | 138 BC patients, 89 controls | RT-PCR | Circulating CAIX mRNA levels: SN = 91%, SP = 72% | Diagnosis of UC | Malentacchi et al.96 |
| Urine mRNA | 181 NMIBC patients | Xpert(R) Bladder Cancer Monitor | Recurrence: SN = 73.7%, SP = 79.6% | Recurrence prediction of NMIBC | Elsawy et al.97 |
| Urine miRNA | 111 BC patients, 25 controls | RT-PCR | miR-200: SN = 62.2%, SP = 100%; miR-145: SN = 78.4%, SP = 91.7%; miR-21: SN = 83.8%, SP = 100% | Diagnosis of BC | Mamdouh et al.103 |
| Urine miRNA | 6 UC patients, 3 controls | TEM, ELISA, RT-PCR, microarray analysis | miR-21-5p: SN = 75%, SP = 95.8% | Diagnosis of UC | Matsuzaki et al.104 |
| Urine miRNA | 10 BC patients, 10 controls | NGS, RT-PCR | NA | Diagnosis of BC | Lin et al.105 |
| Urine microRNA | 8 BC patients, 4 controls | RT-PCR | hsa-miR-1255b-5p: SN = 85%, SP = 68.4% | Diagnosis and surveillance of BC | Tölle et al.106 |
| Urine miRNA and urine protein | 34 BC patients, 20 cystitis, 19 controls | Telomerase extension reaction, RCA | NA | Diagnosis of BC, MIBC and NMIBC | Duan et al. 107 |
| Urine miRNA and SERS | 66 BC patients, 50 controls | NGS, RT-PCR | miRNA: AUC = 0.89; miRNA and SERS: AUC = 0.95 | Diagnosis of BC | Moisoiu et al.108 |
| Urine lncRNA | 230 BC patients, 230 controls | Microarray analysis, RT-PCR | Two lncRNA panel: overall AUC = 0.885; Ta: AUC = 0.843; T1: AUC = 0.867; T2–T4: AUC = 0.923 | Diagnosis of BC and prognosis of NMIBC | Du et al.118 |
| Urine circRNA | 18 BC patients, 14 controls | RT-PCR | NA | Prognosis and metastasis prediction of BC | Chen et al.128 |
| Urine circRNA | 116 BC patients, 30 controls | Microarray analysis, RT-PCR | NA | Diagnosis and prognosis of BC | Song et al.131 |
| Exosomes | |||||
| Urine exosomal DNA | 6 BC patients | RT-PCR, Sanger sequencing, WES | NA | Diagnosis of BC | Zhou et al.137 |
| Urine exosomal miRNAs | 12 BC patients, 4 healthy controls | TEM, Western blot, RT-PCR, WGS | miR-93-5p: SN = 74.1%, SP = 90.2%; miR-516a-5p: SN = 72.9%, SP = 89.9%; miR-93-5p with miR-516a-5p: SN = 85.2%, SP = 82.4% | Diagnosis of BC | Lin et al.138 |
| Urine exosomal mRNA | 168 BC patients, 90 controls | TEM, FCEM, RT-PCR | CA9 mRNA: SN = 85.18%, SP = 83.15% | Diagnosis of BC | Wen et al.139 |
| Extrachromosomal circular DNAs | |||||
| ucf-eccDNA | 21 CKD patients, 28 controls | Circle-Seq | NA | Diagnosis of urogenital disorders | Lv et al.147 |
| Microbiome | |||||
| Urine microbiome DNA | 122 BC patients, 29 controls | 16S rRNA gene sequencing | NA | Diagnosis of BC | Oresta et al.155 |
| Urine microbiome DNA | 31 BC patients, 18 non-neoplastic controls | 16S rRNA gene sequencing | NA | Diagnosis of BC | Wu et al.156 |
- Abbreviations: AMH, asymptomatic microscopic haematuria; AUC, area under curve; circRNA, circular RNA; CKD, chronic kidney disease; ddPCR, droplet digital polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; FCEM, flow cytometry of exosome marker; HG, high grade; HIP/PAP, hepatocarcinoma-intestine pancreas/pancreatitis-associated protein; LG, low grade; lncRNA, long non-coding RNA; MIBC, muscle-invasive bladder cancer; mRNA, messenger RNA; NGS, next-generation sequencing; NMIBC, non-muscle invasive bladder cancer; NMP, nuclear matrix protein; RCA, rolling circle amplification; RT-PCR, real-time polymerase chain reaction; SERS, surface-enhanced Raman spectroscopy; SN, sensitivity; SP, specificity; TEM, transmission electron microscopy; UBC, urothelial bladder carcinoma; UC, urothelial carcinoma; uCAPP-seq, urinary cancer personalised profiling by deep sequencing; ucfDNA, urinary cell-free DNA; ucf-eccDNA, urinary cell-free extrachromosomal circular DNA; uctDNA, urinary circulating tumour DNA; UroCAD, urine-exfoliated cells copy number aberration detector; utMEMA, a method detect urine tumour DNA methylation at multiple regions by MassARRAY; WES, whole exome sequencing; WGS, whole-genome sequencing.
3 CONSIDERATIONS FOR DEVELOPING URINE-BASED BIOMARKERS
In this section, we highlight a few confounding factors that could affect the laboratory test of urine-based biomarkers.
3.1 Urine collection
Urine samples from BC patients consist of a large amount of tumour-derived DNA/RNA. Tumour-derived DNA and RNA can be isolated from full void urine samples, urine pellets and urine supernatants.14 And urinary cell-free DNA (ucfDNA) is characterised as highly fragmented and ranges in size between 80 and 110 bp, which can be used as a liquid biomarker for BC diagnosis and monitoring.15, 16 To ensure the diagnostic accuracy of urine-based liquid biopsy, urine collection is the primary step towards improving the clinical efficacy of diagnosis. Standardisation of the urine collection process is key to ensure the quality of samples and prevent nucleic acid (NA) degradation.15, 17 Operation procedures, collection time and sterile conditions are the main aspects that should be standardised.17 Based on clinical aims and objectives, urine sample processing could be adjusted accordingly. During the urine collection, the quality of the urine sample is affected by several factors. The levels of urinary NA vary at different urine collection time. For instance, the DNA amount in morning-void urine samples is generally higher than that in mid-stage urine, however, morning-void urine contains other debris and fewer impurities.18 In addition, morning-void urine can be contaminated by physiological secretion and organisms of the urethra, genital and other urinary track.19 Overall, high-quality urine samples should be collected by standardising processes to reduce the influence of physiological secretions.
3.2 Urine preservation
After collection, urine should be processed and analysed quickly to avoid the degradation of biomolecules. DNA and RNA in urine samples can be degraded by metal-dependent nucleases, such as deoxyribonuclease I.20 To prevent the degradation of NA, ethylenediaminetetraacetic acid (EDTA) could be added into the urine to reduce the undesirable effects of nucleases.20 In addition, temperature is another important factor for urine preservation. If the urine sample cannot be processed within 2 hours, it should be preserved at –20°C or –80°C for further processing. To avoid NA degradation, urine-conservation media, which includes phosphate-buffered saline, chelating agent and bovine serum albumin, can be used to preserve the potential biomarkers.18 In Table 2, we summarise the available preservatives for DNA, RNA and other biomarkers with suitable application conditions.
| Biomarker | Preservative | Functions/comments |
|---|---|---|
| DNA | EDTA, bovine serum albumin | Chelating agent, inactivated nuclease; overnight storage at 4°C or –80°C |
| Urine conditioning buffer (ZYMO Research) | Commercially available | |
| Urine preservative (Norgen Biotek) | ||
| Cell-free DNA urine preserve (Streck) | ||
| RNA | Guanidine thiocyanate-based buffer | Recovery rate well; whole urine |
| Urine conditioning buffer (ZYMO Research) | Commercially available | |
| Protein | Methylbenzene | Suitable for urine protein qualitative and quantitative analysis |
| Boric acid | Inhibit bacteria in 24 h |
3.3 Differences between urine pellets and supernatant
U cfDNA or other biomarkers exist in both urine pellets (after centrifugation) and urine supernatant, and urine fractions show different sensitivities for different biomarkers. Compared with urine pellets, urine supernatant is more sensitive in detecting microsatellite markers.21 The ucfDNA from urine supernatant also represents a higher genome load compared with urine pellets, which means higher genomic complexity.22 The diagnostic effect of urine pellets also provides insights into detecting DNA methylation. Hentschel et al.23 demonstrated that DNA methylation markers in urine pellets are highly consistent with those in paired tumour samples, especially compared with urine supernatant or full void urine. In addition, urine pellets contain more particles of tumour cells that fall off into the urine, so the pellet DNA is more tumour specific and has a higher correlation with tumours than urine supernatant. In addition, detecting tumour DNA from urine pellets is cost-effective and less laborious. These features make urine pellets as a more desirable source for urine-based liquid biopsy than urine supernatant.
4 THE PRESENCE OF GENOMIC DNA IN URINE OF BC PATIENTS
4.1 Cell-free DNA and circulating tumour DNA
Both cell-free DNA (cfDNA) and circulating tumour DNA (ctDNA) can be detected in urine from BC patients. The cfDNA is a byproduct derived from the apoptosis and necrosis of normal cells and/or cancer cells.24, 25 Currently, plasma cfDNA is widely explored for its potential in tumour detection. ucfDNAs also gain attention due to their close contact with lesions of urinary disease and are therefore considered potential urine biomarkers for BC.26, 27 Recent clinical research found that ucfDNA can monitor the recurrence and progression of NMIBC. Xu et al.28 enrolled 103 patients with NMIBC (pTa-pT1) and measured the IQGAP3/BMP4 expression ratios in ucfDNA by real-time quantitative polymerase chain reaction (RT-qPCR). They found that the NMIBC patients with high IQGAP3/BMP4 expression ratio had a worse prognosis (53.4%) and recurrence (28.2%). This research proves that the IQGAP3/BMP4 ratio in ucfDNA might be an effective liquid biomarker for the diagnosis and recurrence prediction in patients with NMIBC.
ctDNA is a small part of cfDNA, which is identified by carrying tumour-specific genomic alterations.29 Because of incomplete digestion and clearance of phagocytes, it represents random fragments.30 Generally, ctDNA is shorter than cfDNA, ranging from 50 to 150 bp.24, 31, 32 ctDNA is rich in tumour-specific mutations, copy number changes and altered DNA methylation status.33 Recently, urinary ctDNA is increasingly used to diagnose and monitor BC.34 Birkenkamp-Demtröder et al.35 analysed urinary ctDNA from 12 patients with NMIBC (six with recurrence and six with progression to MIBC), and three methods were used to identify genomic alterations in tumour (whole-genome sequencing [WGS], whole-exome sequencing [WES] and mate-pair sequencing). They found that in most patient samples, the level of urinary ctDNA was clearly higher than that in plasma. Moreover, the level of urinary ctDNA differs among different stages of BC.
4.2 Urinary DNA isolation methods
Due to the low concentration of ucfDNA, it is essential to ensure the extraction quality by processing urine samples in an accurate way.36 Different methods have been developed for DNA purification from urine samples, such as organic solvent dissolution, column-based method and magnetic beads capture.37 The organic solvent dissolution method refers to dissolving DNA into an organic solvent (typically chloroform). However, this method is genotoxic, time-consuming and hard to standardise. Column-based method and magnetic beads capture are common methods with relatively high recovery, and therefore widely used.38 Oreskovic et al.15 found that the DNA recovery rate by the silica spin column method is from 30% to 75% from urine, while the recovery amount of magnetic beads capture could be up to 73%–84%. Notably, the column-based method and magnetic beads capture are both efficient in extracting DNA fragment with the short length.15 They also gain the advantages of recovering low-concentration short DNA fragments, being tolerant to different urine environments, and get resistance to PCR inhibition.15 Compared with the column-based method, magnetic beads capture has higher popularity in research and clinical applications due to the availability of automatic operation and higher recovery.39 Based on different research aims, the isolation methods should be adjusted properly.
5 TECHNIQUES FOR EFFICIENT DETECTION OF URINARY TUMOUR DNA
In this section, we present an overview of methods used for the detection of urinary tumour DNA.
5.1 Droplet digital PCR
ctDNA is a rich biomarker of the genetic information from tumour cells. However, in urine, it is difficult to acquire enough tumour features from a scarce amount of urinary DNA.40 Thus, a highly sensitive technique is needed for the detection of urinary ctDNA. Droplet-based digital PCR (ddPCR) is an efficient method based on partitioning. It divides cfDNA into each independent aqueous droplet and detects specific genomic information (such as somatic mutations) and small residual lesions.41 Hayashi et al.42 collected 202 UBC urine sample cohorts and used ddPCR to detect cfDNA mutations, which targets TERT and FGFR3 genes. In the test cohort, the sensitivity of urinary cfDNA in the diagnosis of BC was 68.9%, and the specificity was 100%. There are several sophisticated platforms for commercial ddPCR, such as Bio-Rad QX200TM Droplet Digital TM Systems (Bio-Rad Laboratories), RainDance Technologies RainDrop Digital PCR and Stilla Technologies Naica Platform.41, 43 Taken together, ddPCR is an ideal approach to analyse cfDNA or ctDNA in urine, and further predict UBC recrudescence. The ddPCR has obvious advantages in single molecular detection and is frequently cooperative and applied with next-generation sequencing (NGS).
5.2 Next-generation sequencing
NGS is a high-throughput and sensitive method that can acquire millions of sequencing reads at the same time.44 NGS can be classified into WGS, WES and clinical exome sequencing.45 WGS is a common method to analyse ucfDNA, it can widely capture different genomic changes, including somatic mutations and structural variations.46 Chauhan et al.47 detected ucfDNA by applying urine cancer personalised profiling by deep sequencing (uCAPP-seq) in 42 MIBC patients and 15 control cohorts. uCAPP-seq is similar to NGS and combines cancer personalised profiling by deep sequencing (CAPP-seq) and integrated digital error suppression.48 Recently, uCAPP-seq was developed as a novel urine-based approach for early detection and surveillance of NMIBC.49 Surprisingly, Chauhan et al.47 found that ufDNA minimal residual disease (MRD) detection is highly correlated with pathologic complete response, and the sensitivity and specificity are both 81%. uCAPP-seq can be used as a sensitive method to detect ucfDNA as evidence for choosing radical cystectomy. In 2020, another study used the low-coverage WGS technology (LC-WGS) to detect urine-exfoliated cell DNA.50 This study recruited 219 participants, including 137 urothelial carcinoma (UC) patients and 82 health control cohorts. The sensitivity and specificity of LC-WGS were 82.5% and 96.9%, respectively.50 This result demonstrated that WGS is an available method for the non-invasive detection of urine cfDNA. Traditional NGS is limited to detecting low-frequency mutations. Meanwhile, sequencing depth and wide detecting targets cannot be achieved at the same time, which can be further improved by combining ddPCR and NGS.
5.3 Concentration and integrity
Concentration and integrity are two main typical features of ucfDNA. Brisuda et al.51 found the concentration of ucfDNA can be used as a biomarker to diagnose BC. In this study, 100 volunteers were divided into three groups: (1) 66 BC patients, (2) 23 healthy volunteers and (3) 11 patients with benign urological disorders. Through RT-qPCR, Brisuda et al. found that the concentration of ucfDNA showed a non-negligible difference between the BC group and the control. The diagnosing specificity is much higher than the sensitivity, which is 91.2% and 12%, respectively. They concluded that the concentration of ucfDNA is a potential non-invasive and high-specificity biomarker for the diagnosis of BC, and can be combined with other clinical methods to improve the sensitivity. However, as discussed above, the concentration of ucfDNA can be affected by storage, extraction, sampling time, etc. Cautions of confounding factors should be taken into consideration when using ucfDNA concentration as a biomarker for BC diagnosis.
ucfDNA can also be recognised by its integrity. High molecular weight ucfDNA is defined as a DNA fragment longer than 1000 bp, and it is mainly derived from necrotic cells along the urogenital tract, while low molecular weight ucfDNA is considered as 10–400 bp fragments from cell apoptosis.52 Normally, low molecular weight ucfDNA exists in healthy individuals, while cancer patients mainly contain high molecular weight ucfDNA from tumour cells. However, the accuracy of concentration and integrity is dependent on extraction procedures, which need future validation from large cohorts. Further, BC is a urinary system disease with shed cells in the urine, and from these shed cells, the integrity of ucfDNA could be a marker for monitoring BC development, progression and recurrence in a non-invasive way.53
5.4 Somatic mutation/microsatellite analysis
Somatic mutation is a hallmark of the cancer genome. BC harbours an extremely higher mutation burden than many other cancer types.54 A previous study has explored whether this high mutation of BC can be used as a liquid biopsy way for early screening. In this study, authors recruited 12 BC patients and analysed the preoperative and postoperative urine samples of 11 patients. Indeed, depending on the presence of somatic mutations in BC-related genes, the detection sensitivity of early screening can be increased to 91%.55 These data reveal a rich mutational landscape in BC, and based on these features, somatic mutation can be developed into biomarkers, and become one of the promising strategies for the treatment of BC in the future. ctDNA from urine also has the ability to detect MRD in MIBC for diagnosis. In Chauhan et al.’s research,47 they tested 72 urine samples (48 from BC patients after proceeding radical cystectomy and 27 from the healthy group) by CAPP-seq. They found that urine ctDNA MRD detection was positively correlated with patients without pathologic complete response (sensitivity of 81% and specificity of 81%). And urine ctDNA MRD-positive patients were also related to worse prognosis. Thus, urine ctDNA is an attractive biomarker for diagnosis and personalised treatment.
In addition, based on the copy number variation from cell pellets, new techniques can also be developed with high diagnostic sensitivity. In 2020, Zeng et al.50 established a novel diagnosis model with a customised bioinformatics workflow named Urine Exfoliated Cells Copy Number Aberration Detector (UroCAD), and analysed urine pellet samples from 126 patients and 64 healthy individuals by UroCAD. They found that ucfDNA from the urine pellets has superior sensitivity and specificity in diagnosing UC than traditional cystoscopy.50
Another genomic instability event is microsatellite instability (MSI). Microsatellites are short tandem repeats in genes, when their length changes because of replication errors (such as insertion or deletion), it is called MSI.56 Schneider et al.57 had identified urine microsatellite DNA as BC biomarkers in the early stage combined with fluorescent PCR. The cohort included 103 different stages BC patients, 47 cases of other malignancies, 7 cases of benign inflammatory urothelial diseases and 26 controls without malignant diseases. They observed an overall sensitivity of 84% in detecting BC. There is still a problem that MSI is not enough to be detected from urine in late stages, which requires further development.58
6 EPIGENETIC FEATURES IN BC
Two important categories of epigenetic features, including DNA methylation and histone modifications, can be used as biomarkers for tumour diagnosis and monitoring. Here, we provide the current progress in detecting epigenetic biomarkers from the urine of BC patients.
6.1 Methylation
DNA methylation is a typical form of epigenetic modification that influences genome structure, integrity and gene expression.59 Hypermethylation of the CpG island in promoter regions could lead to downregulated expression of tumour suppressor genes, which are often involved in initiating carcinogenesis process. Therefore, the ‘methylation pattern’ of ctDNA has been the most thoroughly studied hotspot for cancer research in recent years, and recognition of distinct methylation patterns may serve as discriminatory tools for the detection and diagnosis of BC. Multiple gene sites in BC are hypermethylated, including Reprimo (RPRM), prostaglandin-endoperoxide synthase 2 (PTGS2), glutathione stransferase pi 1 (GSTP1) and APC regulator of the Wnt signalling pathway. By performing methylation‑specific RT-qPCR on 477 urine samples (137 BCs, 202 control samples, 28 benign BCs and other related urinary cancer samples), Deng et al.60 found that the methylation level of DMRTA2 shows higher sensitivity in detecting early stages of BC (94.1% for T1, 96.4% for T2, 77.8% for T3 and 71.4% for T4). Moreover, the methylation level of DMRTA2 can also distinguish BC from other interfering urinary cancer and benign BC. However, one methylation marker test is not sensitive for diagnosing Ta, T3 and T4 stages of BCs, which suggests using a combination of different markers. Compared with non-malignant control urines, significant methylation gene combination, such as CyclinD2 and APC, were detected in the urine of malignant BC with high sensitivity and specificity.61 Using quantitative methylation-specific PCR, Bosschieter et al.62 analysed the methylation level of 14 genes in the urine of 72 BC patients and 75 healthy controls. They identified a two-gene panel that could detect BC with high accuracy. Hentschel et al.23 also indicated the correlation between DNA methylation and BC. A study by Renard et al.63 designed a real-time methylation-specific polymerase chain reaction assay for the detection of specific methylation markers, NID2, and the Twist Family BHLH Transcription Factor 1 (TWIST1). The study reported that this two-gene panel had 90% sensitivity and 93% specificity for distinguishing primary BC patients from controls. In addition, Chen et al.64 designed a urine-based diagnosis model with two CpG methylation markers, and it has high-diagnostic sensitivity and specificity. This model can also diagnose early-stage BC, minimal residual and recurrence conditions, which has higher sensitivity than traditional diagnosis methods.64 By performing quantitative methylation-specific PCR with 109 BC patients' urine samples and 100 control samples, Hentschel et al.65 found that GHSR/MAL marker panel is sensitive for BC diagnosis (81% sensitivity and 93% specificity). In particular, it has promising potential for late-stage BC diagnosis and classification of primary and recurrent cases. In addition, by analysing the methylation information of urine pellets from a large cohort, van der Heijden et al.66 developed a three-gene (CFTR, SALL3 and TWIST) methylation classifier. They also demonstrated that in a real clinical scenario (PFBC), combining the three-gene panel and the urine cytology had a high sensitivity predictive value. Su et al.67 analysed the DNA methylation levels of HOXA9, SOX1, NPY, IRAK3, ZO2 and L1-MET in the urine sediments from BC patients and healthy individuals. They found that SOX-1, IRAK3 and Li-MET gene methylation status in urine sediment had a higher recurrence predictivity than cystoscopy and urine cytology.
DNA methylation also improves the sensitivity of other BC diagnosis methods, such as cystoscopy. Fantony et al.68 enrolled 172 patients (63% for NMIBC and 37% for haematuria) and performed a prospective multi-institutional study to evaluate the efficiency of the urine-based TWIST1/NID2-based DNA methylation assay. They found that adding a DNA methylation test to urine cytology will improve the diagnostic performance of BC. Methylation can also be combined with mutation sites as an effective detection method. Roperch and coworkers69 showed that combined analysis of DNA methylation markers (HS3ST2, SEPTIN9 and SLIT2 genes) on urine and FGFR3 mutation status provide a highly accurate non-invasive test for population screening. DNA methylation is already considered a liquid biopsy method to diagnose BC at an early stage and as a potential biomarker. However, gender-related factors and unrelated cells (such as leucocytes) can influence the background of unmethylated values.65 Its sensitivity and specificity need to be further verified in a large cohort, but it might become a conventional method for cancer screening with further research.
6.2 Histone modifications
Histone modification is another key feature of epigenetic marks, which includes phosphorylation, acetylation and sulphonation.70 Histone acetylation can reduce the connection between nucleosomal DNA and histone by neutralising the positive charge of the lysine, making chromatin structure relaxation, and raising DNA accessibility. Generally, transcriptional activity is affected by acetylation level, increased by high acetylation and decreased by low acetylation. Acetylation is a dynamic process regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Therefore, the balance of HATs and HDACs influences transcriptional activity, which is one of the major regulators in epigenetics.71 Lucca et al.72 collected and analysed 227 urine RNA samples from 227 patients with asymptomatic microscopic haematuria (AMH), and 18 random samples were chosen to proceed with gene array analysis. The results showed a significant difference in the expression of HDAC9, HDAC3, tRNA-methyltransferase 1 (TRDMT1) and DNA methyltransferase 1 (DNMT1) among the BC group and controls. To verify this result, the research team tested the remaining 209 samples through the RT-PCR method. Consistent with the analysis above, all BC samples showed high expression levels of HDAC9, HDAC3, TRDMT1 and DNMT1. Collectively, histone modification in urine could reflect the characteristics of BC, and it is necessary for further confirmation in a large cohort.
7 URINE PROTEIN MARKERS IN BC
Proteomic analyses have opened a new horizon for cancer biomarker discovery. Proteins are versatile macromolecules that directly participate in biological functions.73 Analysing the corresponding changes in the tumour-specific proteins is essential in revealing the molecular mechanism of tumourigenesis and development. Matrix metalloproteinase-23 (MMP23B), hepatocarcinoma-intestine pancreas/pancreatitis-associated protein (HIP/PAP), nuclear matrix protein (NMP) and cytokeratin are typical urinary proteins that can be applied in the diagnosis and surveillance of BC.74-76 According to the Allione et al.’s research,77 urinary MMP23B can be used to classify BC patients. They analysed 59 BC urine samples and 57 corresponding controls by enzyme-linked immunosorbent assay (ELISA), and the results revealed that urinary MMP23B was highly correlated with tumour grading. In addition, Nitta et al.74 found that HIP/PAP was highly expressed in BC tumour tissues. An ELISA demonstrated that the level of urinary HIP/PAP could be used to predict the risks of progression and recurrence of BC. NMP is a protein family that constructs nuclear structure, and it is involved in DNA replication and regulation of gene expression. In cancer patients, NMP members are overexpressed and released into urine through cell apoptosis. NMP22 is one of the biomarkers used for the diagnosis or surveillance of BC with Food and Drug Administration approval.78 The laboratory tests for NMP22 are mainly the ELISA test and the BladderCheck test. ELISA is a quantitative test, while the BladderCheck is a qualitative point of care test.79 One study found that detecting the abundance of urinary NMP22 has a sensitivity (63.5%) and specificity (75%) in BC.80 Another study has proved that the concentration of urinary NMP22 can be used as a biomarker to monitor the recurrence of BC.81 These studies have shown that NMP22 has a high specificity in detecting BC, however, it suffers from insufficient sensitivity, especially in low-grade BC, and is highly susceptible to false positives caused by factors including infection, uroliths, inflammation, haematuria, etc.82 Therefore, its clinical application value in BC remains to be explored. The combination of NMP22 and urine laminin-γ2 monomer (Ln-γ2m) shows great ability in detecting NMIBC. By analysing 297 samples (111 NMIBC, 136 benign genitourinary disease and 50 controls) with chemiluminescent immunoassay, Ln-γ2m/uCRN (Ln-γ2m normalised by urine creatinine) is higher expressed in NMIBC, compared with benign diseases and healthy people.83 The diagnostic effect of the combination of NMP22 and Ln-γ2m/uCRN in NMIBC is more ideal than NMP22 or Ln-γ2m/uCRN independently.
Cytokeratin, one of the intermediate filament proteins of the epithelial cytoskeleton, is another commonly used biomarker for BC detection.27 After epithelial cell death, cytokeratin is released into urine, and its concentration can be detected to predict the absence or presence of BC.84 It has been demonstrated that cytokeratin 8, 18, 19 and 20 are significantly associated with the progression BC.85-87 The abundance of cytokeratin was detected by using the mass spectrometry, and its results can be easily affected by other high-abundance proteins.88 However, there are a large number of normal cells and cell metabolic components in urine, which may influence the concentration of cytokeratin and other protein biomarkers. Consequently, protein biomarkers are not widely used, especially when compared with other urinary biomarkers.
8 URINE RNA BIOMARKERS IN BC
Various RNAs exist in human urine, including mRNA, miRNAs, lncRNAs, circRNAs and tRFs. During tumour progression, different RNA expression and modification levels are dynamic. To a certain extent, urinary RNA can be used as a biomarker for the surveillance of the diagnosis and prognosis of cancer. Currently, some RNA molecular biomarkers have been identified in urine biopsy studies of BC.89-91
8.1 mRNAs
Several studies have focused on the clinical application of urinary mRNA in the detection of BC. By using TaqMan-based RT-PCR, Hanke et al.92 found that a large amount of cell-free RNA exists in the whole urine, which figured out the ideal source for mRNA isolation. Multiple mRNA types are considered potential BC biomarkers with ideal diagnostic effects. Kim et al.93 analysed the levels of IQGAP3 and TOP2A from the urine samples of 92 BC patients and 120 controls. Compared to normal specimens, urinary IQGAP3 was significantly higher in BC patients. Further analysis showed that IQGAP3 can discriminate between BC patients and non-cancer patients with haematuria symptoms. In addition, high level of UBE2C was also clearly related to BC (82.5% sensitivity and 76.2% specificity).94 Besides the high level of mRNA biomarkers, downregulated mRNA levels also provide insights into BC status. Zhang et al.95 measured the level of N-Myc downstream-regulated gene 2 (NDRG2) in urine from 97 healthy donors and 127 BC patients. Their results showed that the expression level of NDRG2 was clearly downregulated in BC patients. It can also be used to identify BC patients with different stages. In addition, by performing RT-PCR on RNA from urine sediments of BC patients, Malentacchi et al.96 found that mRNA in urine was also consistent with corresponding tissue detection (80% concordance). Elsawy et al.97 designed an mRNA-based Urine Test panel (Xpert Bladder Cancer Monitor), which target on ABL1, ANXA10, UPK1B, CRH and IGF2. By analysing 181 NMIBC patients, this mRNA panel shows great potential in diagnosing recurrence (sensitivity 73.7%), which is more sensitive than urine cytology (sensitivity 47%).
8.2 miRNAs
miRNA is a class of short non-coding RNA, and the length of miRNA is around 18–24 nucleotides.98 miRNA regulates cancer proliferation and metastasis by complementarily pairing with mRNA. It also mediates and silences gene expression at the post-transcriptional level.99 Tumour-derived miRNAs could be released from exosomes to urine or bloodstream.100 Some studies have shown that the expression level of some miRNAs could be an independent biomarker for the detection and monitoring of BC.101 Some miRNAs act as oncogenes in BC development.102 Mamdouh et al.103 analysed the expression level of miR-200, miR-145 and miR-21 in 136 urine samples (111 from BC patients and 25 from healthy people) through RT-PCR. They identified that the expression level of miRNA in the urine is consistent with that in tissues. A small cohort study also showed that miR-21-5p can be used to diagnose BC by performing microarray. Matsuzaki et al.104 analysed six UC patients and three controls by miRNA profiling and identified miR-21-5p overexpressed in UC patients which have negative urine cytology (75% sensitivity and 95.8% specificity). These results collectively demonstrated the possible potential for urinary miRNA in BC diagnosis. According to Lin and Tsai's research,105 miR-146a-5p, miR-149-5p and miR-423-5p are highly presented in BC patients by analysing urine samples of 70 BC patients and 90 controls. The levels of urinary miR-618 and miR-1255b-5p in MIBC patients were significantly higher than those in the healthy group; the sensitivity and specificity of miR-1255b-5p were 68% and 85%, respectively, in the diagnosis of MIBC.106 Recently, co-detection of miRNA with other biomolecules has improved the sensitivity and specificity of BC diagnosis and even can distinguish BC subtypes. Duan et al.107 found that the combination of telomerase and urinary miR-21 levels has the potential to detect cystitis and BC. This detection method can also distinguish MIBC and NMIBC, which can be used as a piece of preclinical evidence to choose a suitable clinical therapy strategy.107 miRNA detection can also proceed with other techniques. Combined with surface-enhanced Raman spectroscopy, miRNA detection has high classification accuracy for luminal and basal BC.108 Meanwhile, NGS can also be used to capture urine miRNA profiles, which has better subtype judgments in BC patients.109 Through genome-wide urinary miRNA profile analysis, increasing levels of urinary miR-31-5p, miR-191-5p and miR-93-5p in BC patients are found comparable with that in the healthy group. However, the results from NGS and RT-PCR only partially agree, and further steps are needed to verify the accuracy and reliability of the results from both methods.110
8.3 lncRNAs
lncRNAs are a class of non-coding RNAs that exist in eukaryotes with a length of more than 200 nucleotides.111, 112 They can regulate gene expression and epigenetic modifications. Dysregulation of lncRNAs is commonly found in various diseases and cancers.113 A broad range of cellular events including immunomodulation, angiogenesis, invasiveness, migration and differentiation are reported to be influenced by lncRNAs.114 By highlighting the role of lncRNAs in the physiological progression of carcinogenesis, recent studies have explored their clinical utility in cancer diagnosis and prognosis.115 Many urinary lncRNAs are derived from exosomes without the disruption of endogenous RNAase activity.116 RT-qPCR is considered as a robust tool with high accuracy for detecting urinary lncRNAs in liquid biopsy.117 Using microarray assay, Du et al.118 constructed a two-lncRNA (GAS5 and uc004cox.4) panel for the detection of BC with an AUC value of 0.885. Meanwhile, uc004cox.4 could provide prognostic information for BC, and a higher expression level of uc004cox.4 was associated with poorer OS and a higher recurrence rate.
8.4 tRFs
tRNA is a common type of RNA that exists in all living organisms. tRFs are usually derived from mature tRNA and tRNA precursors, with a length of 13–48 nucleotides.119 tRFs can be categorised into tRF-1, tRF-3, and tRF-5 based on different splicing sites.33 Therefore, some prior studies explored using tRFs in body fluids as new biomarkers for cancer diagnosis. For instance, 5′-tRF-LysCTT in plasma is significantly elevated in BC patients with early progression and poor prognosis.120 Growing evidence suggested that tRFs regulate gene expression and other biological activities, and they have also been detected in the urine of different cancers.121 The abundance of tRFs in urine is also one of the highest among different body fluids.121 However, only a few studies have indicated the application of urinary tRFs in BC diagnosis, and more research is necessary in the future to demonstrate the effect of tRFs in BC urinary biopsy.
8.5 circRNA
circRNA is a covalent-linking single RNA loop without 3′ polyadenylated tails and 5′ caps.122 The unique circular structure of circRNAs enables them to have higher stability than linear RNA, which causes more chances for circRNA to participate in physiological regulation.123 Besides acting as a translation template, recent studies have explained that circRNAs can combine with miRNAs to inhibit miRNA function.124, 125 Emerging evidence shows that specific circRNAs participate in BC initiation and progression, and are considered as potential biomarkers for BC urine biopsy.126-129 Chen et al.128 collected 18 urine samples from BC patients and 14 corresponding urine samples from healthy people to analyse the relationship between the circRNA in urine and BC development. After extracting circRNA from exosomes and analysing it by RT-PCR, they found that circPRMT5 is also highly correlated with BC lymph node metastasis and tumour progression. Moreover, compared with other cancer diagnosing methods, urinary circRNAs show higher accuracy in predicting cancer progression.130 According to Song et al.’s study,131 hsa_circ_0137439 is an outstanding circRNA marker in BC prognosis. After performing RT-PCR on 116 BC urine samples and 30 controls, they found that hsa_circ_0137439 was upregulated in BC samples, which shows great potential in distinguishing MIBC from NMIBC. However, insufficient studies have applied circRNA as a urine biopsy biomarker in a large cohort, and its wide application for circRNA in urine biopsy needs to be further confirmed.
9 EXOSOMES IN URINE OF BC
Exosomes are 30–150 nm vesicles that contain various active components, such as proteins, NAs, carbohydrates and lipids.132 Exosomes are involved in cellular activities as diverse as modelling the extracellular matrix and transmitting signals and molecules to other cells.133-135 Recently, a few studies reported that BC-specific genes in exosomes are involved in immune activities.136 Based on these findings, urinary exosomes can act as a novel non-invasive biomarker of BC, including detecting somatic variants, mRNA or miRNAs.137, 138 By performing 168 BC urine samples and 90 controls with RT-PCR, the high expression level of CA9 mRNA in urinary exosomes was highly correlated with BC patients (85.28% sensitivity and 83.15% specificity).139 Zhou et al.137 also found that multiple somatic variants (e.g., miRNA-binding regions of the KRAS gene) were found in DNA from urinary exosomes of BC patients. Lin et al.138 utilised urinary exosomes-miRNAs as biomarkers for BC, and they also conducted experimental verification of the mechanism of miR-93-5p in BC. Compared with the healthy group, the expression of miR-93-5p was significantly increased in BC. After further investigation, the author found that miR-93-5p could promote the proliferation and migration of BC cancer cells by inhibiting the expression of BTG2. Therefore, exosome-rich inclusions make it feasible to become a hallmark for the detection of multiple diseases.
10 EXTRACHROMOSOMAL CIRCULAR DNAS
eccDNAs are DNA circles originating from the linear chromosome.140 eccDNAs play an essential role in cancer pathogenesis and are ubiquitously present in living organisms from prokaryotes to eukaryotes.141, 142 The size of eccDNAs varies greatly, from dozens of bases to tens of thousands, but the majority of eccDNA is less than 500 bp.143, 144 Multiple models have been postulated to explain how eccDNA formed in human cells, including the episome model, translocation–deletion–amplification model, breakage–fusion–bridge model and chromothripsis.145, 146 Recently, our previous work along with studies from other groups demonstrated that eccDNA is also present in the plasma and urine samples, and shows some connection with a multitude of diseases.147 For example, Kumar et al.148 found that both cancerous and normal tissues could release eccDNA into the circulation. Besides, the size of plasma eccDNA in cancer patients after surgery is clearly shorter compared to the same patients before surgical resection. Interestingly, by enriching plasma eccDNA from pregnant women, Lo and coworkers149 demonstrated that the foetal-derived eccDNA is generally hypomethylated and shorter compared to maternal-derived eccDNA, and the size of eccDNA was positively correlated with its methylation density. eccDNA is also present in human urine. By developing a new eccDNA purification method from urine supernatant samples, our team found that urinary cell-free eccDNA (ucf-eccDNA) of chronic kidney disease (CKD) patients showed distinct profiles from those of normal individuals. ucf-eccDNA was significantly enriched in CKD patients. A series of ucf-eccDNAs carrying miRNA genes (e.g., miR4508, miR1290, miR2278, miR409 and miR4521) were specifically found in CKD patients.147, 150-153 Owing to its closed circular structure, eccDNA is more resistant to exonuclease digestion than linear DNA. Therefore, they can serve as new biomarkers to provide new insights for liquid biopsy.154
11 URINE MICROBIOME BIOMARKERS IN BC
In addition to these biomolecules, there are microbiomes in the urine. Although there are few studies on the microbiome in BC, some researchers have found that a variety of the urinary microbiome might also be involved in the progression of BC. Oresta et al.155 analysed urinary DNA by 16S rRNA sequencing and found that BC patients have a higher abundance of Veillonella and Corynebacterium, and decreased level of Ruminococcus. The differences in microbiomes in different progression stages of BC are also exacerbating. By analysing DNA isolated from urine pellets, Wu et al.156 found that enrichment of Bacteroides, Porphyrobacter and Herbaspirillum was correlated with a high risk of recurrence and progression in BC. In one study, DNA of urine specimens from UC patients (n = 8) and healthy individuals (n = 6) was sequenced. It was found that Streptococcus was enriched in BC patients.157 Owing to the microbiome diversity being affected by age, the microbiome is hard to be a powerful biomarker of BC.158
12 LIMITATIONS OF URINARY-BASED LIQUID BIOPSY
Urine-based liquid biopsy has several limitations. Firstly, urine sample collection is one of the major factors that could influence the accuracy of liquid biopsy results. Different time-point urine samples from the same individual patient may have different results.159 Secondly, urine samples generally contain a large number of nucleases, which are released from bacteria. Typically, the morning urine samples had both the highest cell content and nucleases, but nuclease has a negative influence on urinary DNA/RNA extraction and therefore affects the test results.160 In order to reduce the effect of nucleases, urine samples must be processed and analysed immediately after collection or stored at 4°C/–20°C for further analysis. The lower storage temperature could inhibit the nuclease activity.37 Thirdly, urine samples contain various components that interfere with liquid biopsy detection. For example, urine contains a large number of normal urothelial cells, which influences the specificity for the detection of tumour cells. In addition, urine also contains a large amount of urea, which is an inhibitor of PCR and therefore affects subsequent analysis and detection.161 Therefore, standardisation of urine processing (e.g., urine sample collection and preservation, DNA/RNA extraction, etc.) is crucial to the accuracy and stability of urine-based biopsy. Another influencing factor of urine-based biopsy is the characteristics of biomarkers themselves. ucfDNA is highly fragmented and ranges from 50 to 100 bp, and the majority of urinary mRNAs are degraded by RNases.162 These features make the information on urinary biomarkers limited and can only be used as auxiliary means of tissue biopsy. If these problems could be overcome, urine will be an ideal clinical source for the screening and monitoring of BC.
13 CONCLUSIONS
Urine-based liquid biopsy is expected to improve the diagnosis, prognosis and treatment of BC patients.163 It has the advanced characteristics of non-invasiveness, high sensitivity, reasonable confidence and low cost, which meet the requirements of early clinical screening. This review provides an overview of the advantages, as well as limitations, of urinary biomarkers and their applications in BC. Meanwhile, more clinical trials on these urinary markers are urgently needed to better understand and explore their clinical translation values.
AUTHOR CONTRIBUTIONS
Dr.Zeng Y., Dr.Wang A., Dr.Lv W., Dr.Wang Q., Dr.Jiang S., Dr.Pan X., and Dr.Wang F. contributed to the preparation and collection of original literatures, and preparation of the figures. Dr.Zeng Y., Dr.Wang A., Dr.Lv W., Dr. Han P., and Dr.Luo Y. contributed to the writing of the original draft. All authors contributed to the editing and critical comments of the manuscript. Dr. Yang H., Dr. Bolund L., Dr. Lin C., Dr. Han P. and Dr.Luo Y. supervised the study. Dr.Zeng Y., Dr.Wang A., Dr.Lv W., Dr. Han P. and Dr.Luo Y. were responsible for the structural designs, scientific quality and writing.
ACKNOWLEDGEMENTS
This work was supported by Qingdao-Europe Advanced Institute for Life Sciences, BGI-Qingdao (BGI-QD). Wei Lv was supported by the China Scholarship Council (CSC).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
The authors received no specific funding for this work.
ETHICAL APPROVAL
Not applicable.
[Correction added on July 15,2023 after first online publication: the Author Contributions, Data Availability Statement and Ethical approval has been updated]
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.




