Phytochemical Profile, Antioxidant, Enzyme Inhibition, Acute Toxicity, In Silico Molecular Docking and Dynamic Analysis of Apis Mellifera Propolis as Antidiabetic Supplement
Corresponding Author
Putri Hawa Syaifie
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Conceptualization (lead), Data curation (equal), Formal analysis (equal), Methodology (equal), Project administration (lead), Visualization (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorDelfritama Ibadillah
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Visualization (equal), Writing - original draft (equal)
Search for more papers by this authorMuhammad Miftah Jauhar
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Biomedical Engineering, Graduate School of Universitas Gadjah Mada, Sleman, 55281 Yogyakarta, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Visualization (equal), Writing - original draft (equal)
Search for more papers by this authorRikania Reninta
Research Center for Applied Botany, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Formal analysis (equal), Validation (equal), Visualization (supporting), Writing - original draft (equal)
Search for more papers by this authorSri Ningsih
Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorDonny Ramadhan
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorAdzani Gaisani Arda
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of Debrecen, Debrecen, H-4032 Hungary
Contribution: Data curation (equal), Formal analysis (equal)
Search for more papers by this authorDhecella Winy Cintya Ningrum
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorNofa Mardia Ningsih Kaswati
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorNurul Taufiqu Rochman
Research Center for Advanced Materials, National Research and Innovation Agency (BRIN), South Tangerang, 15314 Indonesia
Contribution: Funding acquisition (lead), Resources (equal)
Search for more papers by this authorCorresponding Author
Etik Mardliyati
Research Center for Vaccine and Drugs, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Funding acquisition (equal), Resources (equal), Supervision (lead), Validation (equal), Visualization (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Putri Hawa Syaifie
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Conceptualization (lead), Data curation (equal), Formal analysis (equal), Methodology (equal), Project administration (lead), Visualization (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorDelfritama Ibadillah
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Visualization (equal), Writing - original draft (equal)
Search for more papers by this authorMuhammad Miftah Jauhar
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Biomedical Engineering, Graduate School of Universitas Gadjah Mada, Sleman, 55281 Yogyakarta, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Visualization (equal), Writing - original draft (equal)
Search for more papers by this authorRikania Reninta
Research Center for Applied Botany, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Formal analysis (equal), Validation (equal), Visualization (supporting), Writing - original draft (equal)
Search for more papers by this authorSri Ningsih
Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorDonny Ramadhan
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorAdzani Gaisani Arda
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of Debrecen, Debrecen, H-4032 Hungary
Contribution: Data curation (equal), Formal analysis (equal)
Search for more papers by this authorDhecella Winy Cintya Ningrum
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorNofa Mardia Ningsih Kaswati
Center of Excellece Life Sciences, Nano Center Indonesia, Jl. PUSPIPTEK, South Tangerang, 15314 Banten, Indonesia
Contribution: Data curation (equal), Formal analysis (equal), Methodology (equal)
Search for more papers by this authorNurul Taufiqu Rochman
Research Center for Advanced Materials, National Research and Innovation Agency (BRIN), South Tangerang, 15314 Indonesia
Contribution: Funding acquisition (lead), Resources (equal)
Search for more papers by this authorCorresponding Author
Etik Mardliyati
Research Center for Vaccine and Drugs, National Research and Innovation Agency (BRIN), Cibinong, 16911 Indonesia
Contribution: Funding acquisition (equal), Resources (equal), Supervision (lead), Validation (equal), Visualization (equal), Writing - review & editing (equal)
Search for more papers by this authorAbstract
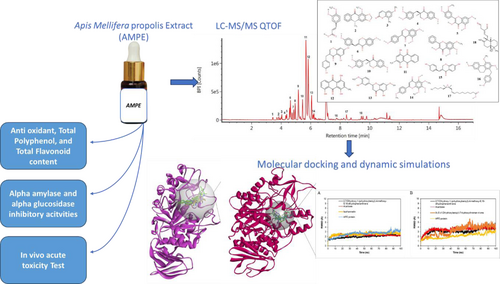
This study aims to identify the phytochemical profile of Apis mellifera propolis and explore the potential of its anti-diabetic activity through inhibition of α-amylase (α-AE), α-glucosidase(α-GE), as well as novel antidiabetic compounds of propolis. Apis mellifera propolis extract (AMPE) exhibited elevated polyphenol 33.26±0.17 (mg GAE/g) and flavonoid (15.45±0.13 mg RE/g). It also indicated moderate strong antioxidant activity (IC50 793.09±1.94 μg/ml). This study found that AMPE displayed promising α-AE and α-GE inhibition through in vitro study. Based on LC–MS/MS screening, 18 unique AMPE compounds were identified, with majorly belonging to anthraquinone and flavonoid compounds. Furthermore, in silico study determined that 8 compounds of AMPE exhibited strong binding to α-AE that specifically interacted with its catalytic residue of ASP197. Moreover, 2 compounds exhibit potential inhibition of α-GE, by interacting with crucial amino acids of ARG315, ASP352, and ASP69. Finally, we suggested that 2,7-Dihydroxy-1-(p-hydroxybenzyl)-4-methoxy-9,10-dihydrophenanthrene and 3(3-(3,4-Dihydroxybenzyl)-7-hydroxychroman-4-one as novel inhibitors of α-AE and α-GE. Notably, these compounds were initially discovered from Apis mellifera propolis in this study. The molecular dynamic analysis confirmed their stable binding with both enzymes over 100 ns simulations. The in vivo acute toxicity assay reveals AMPE as a practically non-toxic product with an LD50 value of 16,050 mg/kg. Therefore, this propolis may serve as a promising natural product for diabetes mellitus treatment.
Graphical Abstract
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Open Research
Data Availability Statement
Data is available in the article.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| cbdv202400433-sup-0001-misc_information.pdf436 KB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1J. B. Cole, J. C. Florez, Nat Rev Nephrol 2020, 16, 377–390.
- 2N. H. Cho, J. E. Shaw, S. Karuranga, Y. Huang, J. D. da Rocha Fernandes, A. W. Ohlrogge, B. Malanda, Diabetes Res. Clin. Pract. 2018, 138, 271–281.
- 3N. J. Morrish, S.-L. Wang, L. K. Stevens, J. H. Fuller, H. Keen, Diabetologia 2001, 44, S14–S21.
- 4O. M. Ogunyemi, G. A. Gyebi, A. Saheed, J. Paul, V. Nwaneri-Chidozie, O. Olorundare, J. Adebayo, M. Koketsu, N. Aljarba, S. Alkahtani, G. E.-S. Batiha, C. O. Olaiya, Front Mol Biosci 2022, 866719, 1–19, DOI 10.3389/fmolb.2022.866719.
- 5J. Wang, H. Wang, Oxid Med Cell Longev 2017, 2017, 1–9.
- 6A. S. Dabhi, N. R. Bhatt, M. J. Shah, Journal of Clinical and Diagnostic Research 2013, 7, 3023.
- 7N. Kaur, V. Kumar, S. K. Nayak, P. Wadhwa, P. Kaur, S. K. Sahu, Chem. Biol. Drug Des. 2021, 98, 539–560.
- 8E. Bonora, Int. J. Clin. Pract. 2007, 61, 19–28.
- 9J. T. Manandhar Shrestha, H. Shrestha, M. Prajapati, A. Karkee, A. Maharjan, Journal of Lumbini Medical College 2017, 5, 34.
10.22502/jlmc.v5i1.126 Google Scholar
- 10J. Ashraf, E. U. Mughal, A. Sadiq, N. Naeem, S. A. Muhammad, T. Qousain, M. N. Zafar, B. A. Khan, M. Anees, J. Mol. Struct. 2020, 1218, 128458.
- 11K. Venkatakrishnan, H.-F. Chiu, C.-K. Wang, J. Funct. Foods 2019, 57, 425–438.
- 12American Diabetes Association, Diabetes Care 2018, 41, S55–S64.
- 13S. Huang, C.-P. Zhang, K. Wang, G. Li, F.-L. Hu, Molecules 2014, 19, 19610–19632.
- 14N. Zabaiou, A. Fouache, A. Trousson, S. Baron, A. Zellagui, M. Lahouel, J.-M. A. Lobaccaro, Chem. Phys. Lipids 2017, 207, 214–222.
- 15M. Franchin, I. A. Freires, J. G. Lazarini, B. D. Nani, M. G. da Cunha, D. F. Colón, S. M. de Alencar, P. L. Rosalen, Eur. J. Med. Chem. 2018, 153, 49–55.
- 16M. Zakerkish, M. Jenabi, N. Zaeemzadeh, A. A. Hemmati, N. Neisi, Sci. Rep. 2019, 9, 7289.
- 17H. R. Sameni, P. Ramhormozi, A. R. Bandegi, A. A. Taherian, M. Mirmohammadkhani, M. Safari, J Diabetes Investig 2016, 7, 506–513.
- 18L. Robles, Pancreat Disord Ther 2013, 1000112, 1–4, DOI 10.4172/2165-7092.1000112.
- 19R. Hossain, C. Quispe, R. A. Khan, A. S. M. Saikat, P. Ray, D. Ongalbek, B. Yeskaliyeva, D. Jain, A. Smeriglio, D. Trombetta, R. Kiani, F. Kobarfard, N. Mojgani, P. Saffarian, S. A. Ayatollahi, C. Sarkar, M. T. Islam, D. Keriman, A. Uçar, M. Martorell, A. Sureda, G. Pintus, M. Butnariu, J. Sharifi-Rad, W. C. Cho, Chin. Med. 2022, 17, 100.
- 20D. Devequi-Nunes, B. A. S. Machado, G. de A. Barreto, J. Rebouças Silva, D. F. da Silva, J. L. C. da Rocha, H. N. Brandão, V. M. Borges, M. A. Umsza-Guez, PLoS One 2018, 13, e0207676.
- 21A. Kurek-Górecka, Ş. Keskin, O. Bobis, R. Felitti, M. Górecki, M. Otręba, J. Stojko, P. Olczyk, S. Kolayli, A. Rzepecka-Stojko, Plants 2022, 11, 1203.
- 22T. M. Kegode, J. L. Bargul, H. O. Mokaya, H. M. G. Lattorff, R. Soc. Open Sci. 2022, 1000112, 1–4, DOI 10.1098/rsos.211214.
- 23L. Svečnjak, Z. Marijanović, P. Okińczyc, P. M. Kuś, I. Jerković, Antioxidants 2020, 337, 1–32, DOI 10.3390/antiox9040337.
- 24Z. Man, Y. Feng, J. Xiao, H. Yang, X. Wu, Front Nutr 2022, 948027, 1–1, DOI 10.3389/fnut.2022.948027.
- 25A. G. Arda, P. H. Syaifie, D. Ramadhan, M. M. Jauhar, D. W. Nugroho, N. M. N. Kaswati, A. Noviyanto, M. Safihtri, N. T. Rochman, D. Andrianto, E. Mardliyati, J Pharm Pharmacogn Res 2024, 12, 264–285.
- 26B. Kumar, J. Devi, A. Dubey, A. Tufail, S. Sharma, Inorg. Chem. Commun. 2024, 159, 111674.
- 27R. Maurus, A. Begum, L. K. Williams, J. R. Fredriksen, R. Zhang, S. G. Withers, G. D. Brayer, Biochemistry 2008, 47, 3332–3344.
- 28K. Yamamoto, H. Miyake, M. Kusunoki, S. Osaki, FEBS J. 2010, 277, 4205–4214.
- 29R. Zrieq, I. Ahmad, M. Snoussi, E. Noumi, M. Iriti, F. D. Algahtani, H. Patel, M. Saeed, M. Tasleem, S. Sulaiman, K. Aouadi, A. Kadri, Int. J. Mol. Sci. 2021, 10693, 1–21, DOI 10.3390/ijms221910693.
- 30C. A. Lipinski, Drug Discov Today Technol 2004, 1, 337–341.
- 31D. F. Veber, S. R. Johnson, H.-Y. Cheng, B. R. Smith, K. W. Ward, K. D. Kopple, J. Med. Chem. 2002, 45, 2615–2623.
- 32P. H. Syaifie, A. H. Harisna, M. A. F. Nasution, A. G. Arda, D. W. Nugroho, M. M. Jauhar, E. Mardliyati, N. N. Maulana, N. T. Rochman, A. Noviyanto, A. J. Banegas-Luna, H. Pérez-Sánchez, Molecules 2022, 3927, 1–3, DOI 10.3390/molecules27133972.
- 33A. J. Owoloye, F. C. Ligali, O. A. Enejoh, A. Z. Musa, O. Aina, E. T. Idowu, K. M. Oyebola, PLoS One 2022, 17, e0268269.
- 34L. Martínez, PLoS One 2015, 10(3), e0119264, DOI 10.1371/journal.pone.0119264.
- 35A. A. Njan, E. O. Olaleye, S. O. Afolabi, I. Anoka-Ayembe, G. A. Gyebi, A. Nyamngee, U. N. Okeke, S. O. Olaoye, F. M. Alabi, O. P. Adeleke, H. D. Ibrahim, Inform Med Unlocked 2023, 39, 101261.
10.1016/j.imu.2023.101261 Google Scholar
- 36I. Aier, P. K. Varadwaj, U. Raj, Sci. Rep. 2016, 6, 1–10.
- 37S. Ahmed, M. Ali, R. Ruma, S. Mahmud, G. Paul, M. Saleh, M. Alshahrani, A. Obaidullah, S. Biswas, M. Rahman, M. Rahman, M. Islam, Molecules 2022, 27, 4526.
- 38M. L. Marcovecchio, M. Lucantoni, F. Chiarelli, Diabetes Technol. Ther. 2011, 13, 389–394.
- 39M. Ghasemi-Dehnoo, H. Amini-Khoei, Z. Lorigooini, M. Rafieian-Kopaei, Asian Pac. J. Trop. Med. 2020, 13, 431–438.
- 40P. Zhang, T. Li, X. Wu, E. C. Nice, C. Huang, Y. Zhang, Front Med 2020, 14, 583–600.
- 41H.-F. Chiu, Y.-C. Han, Y.-C. Shen, O. Golovinskaia, K. Venkatakrishnan, C.-K. Wang, J. Cancer Prev. 2020, 25, 70–78.
- 42A. Ghorbani, R. Rashidi, R. Shafiee-Nick, Biomed. Pharmacother. 2019, 111, 947–957.
- 43U. Gašić, I. Ćirić, T. Pejčić, D. Radenković, V. Djordjević, S. Radulović, Ž. Tešić, Antioxidants 2020, 9, 547.
- 44H. A. El Rabey, M. N. Al-Seeni, A. S. Bakhashwain, Evid.-Based Complement. Altern. Med. 2017, 2017, 1–14.
- 45A. Zeb, J. Food Biochem. 2020, 44(9), e13394, DOI 10.1111/jfbc.13394.
- 46F. Ahmadinejad, S. Geir Møller, M. Hashemzadeh-Chaleshtori, G. Bidkhori, M.-S. Jami, Antioxidants 2017, 6, 51.
- 47J. O. Olugbami, M. A. Gbadegesin, O. A. Odunola, Afr. J. Med. Med. Sci. 2014, 43, 101–109.
- 48C. Sun, C. Zhao, E. C. Guven, P. Paoli, J. Simal-Gandara, K. M. Ramkumar, S. Wang, F. Buleu, A. Pah, V. Turi, G. Damian, S. Dragan, M. Tomas, W. Khan, M. Wang, D. Delmas, M. P. Portillo, P. Dar, L. Chen, J. Xiao, Food Front 2020, 1, 18–44.
10.1002/fft2.15 Google Scholar
- 49N. Asem, N. A. Abdul Gapar, N. H. Abd Hapit, E. A. Omar, J. Apic. Res. 2020, 59, 437–442.
- 50A. E. Z. Hasan, D. Mangunwidjaja, T. C. Sunarti, O. Suparno, A. Setiyono, Emir J Food Agric 2014, 26, 390–398.
- 51E. Barber, M. J. Houghton, G. Williamson, Food 2021, 10, 1939.
- 52C. M. Khoo, in International Encyclopedia of Public Health, Elsevier, 2017, pp. 288–293.
10.1016/B978-0-12-803678-5.00108-9 Google Scholar
- 53H. Laaroussi, P. Ferreira-Santos, Z. Genisheva, M. Bakour, D. Ousaaid, J. A. Teixeira, B. Lyoussi, Food Biosci. 2021, 42, 101160.
- 54A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, C. T. Supuran, Nat. Rev. Drug Discovery 2021, 20, 200–216.
- 55L. Han, C. Fang, R. Zhu, Q. Peng, D. Li, M. Wang, Int. J. Biol. Macromol. 2017, 95, 520–527.
- 56H. Azizian, K. Pedrood, A. Moazzam, Y. Valizadeh, K. Khavaninzadeh, A. Zamani, M. Mohammadi-Khanaposhtani, S. Mojtabavi, M. A. Faramarzi, S. Hosseini, Y. Sarrafi, H. Adibi, B. Larijani, H. Rastegar, M. Mahdavi, Sci. Rep. 2022, 12, 14870.
- 57R. P. P. Neves, P. A. Fernandes, M. J. Ramos, J. Chem. Inf. Model. 2022, 62, 3638–3650.
- 58O. M. Ogunyemi, G. A. Gyebi, A. Saheed, J. Paul, V. Nwaneri-Chidozie, O. Olorundare, J. Adebayo, M. Koketsu, N. Aljarba, S. Alkahtani, G. E.-S. Batiha, C. O. Olaiya, Front. Mol. Biosci. 2022, 866719, DOI 10.3389/fmolb.2022.866719.
- 59Y. Zhao, M. Wang, G. Huang, J. Funct. Foods 2021, 86, 104739.
- 60C. Jiang, Y. Chen, X. Ye, L. Wang, J. Shao, H. Jing, C. Jiang, H. Wang, C. Ma, Int. J. Biol. Macromol. 2021, 172, 503–514.
- 61X. Zheng, H. Chi, S. Ma, L. Zhao, S. Cai, LWT 2023, 178, 114629.
- 62Q. Dong, N. Hu, H. Yue, H. Wang, Molecules 2021, 26, 4566.
- 63J. Lee, E. Jung, J. Lee, S. Kim, S. Huh, Y. Kim, Y. Kim, S. Y. Byun, Y. S. Kim, D. Park, Obesity 2009, 17, 226–232.
- 64N. M. N. Kaswati, M. Bintang, E. Mardliyati, OSR Journal Of Pharmacy and Biological Sciences 2021, 16, 1–12.
- 65S. Forli, R. Huey, M. E. Pique, M. F. Sanner, D. S. Goodsell, A. J. Olson, Nature Protocols 2016, 11, 905–919.
- 66H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, P. E. Bourne, Nucleic Acids Res. 2000, 28, 235–242.
- 67S. Dallakyan, A. J. Olson, Small-molecule library screening by docking with PyRx, 2015, pp. 243–250.
- 68H. Yang, C. Lou, L. Sun, J. Li, Y. Cai, Z. Wang, W. Li, G. Liu, Y. Tang, Bioinformatics 2019, 35, 1067–1069.
- 69X. Li, L. Chen, F. Cheng, Z. Wu, H. Bian, C. Xu, W. Li, G. Liu, X. Shen, Y. Tang, J. Chem. Inf. Model. 2014, 54, 1061–1069.
- 70M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess, E. Lindahl, SoftwareX 2015, 1–2, 19–25.
10.1016/j.softx.2015.06.001 Google Scholar