Inhibition of Putative Ibrutinib Targets Promotes Atrial Fibrillation, Conduction Blocks, and Proarrhythmic Electrocardiogram Indices: A Mendelian Randomization Analysis
Hongxuan Xu, Bingxun Li, and Pinchao Lv contributed equally to this work and share the co-first authorship.
ABSTRACT
Background
The mechanism by which ibrutinib, a Bruton's tyrosine kinase inhibitor, can elevate the risk of arrhythmias is not fully elucidated. In this study, we explored how inhibition of off-target kinases can contribute to this phenomenon.
Methods
We performed a Mendelian randomization analysis to examine the causal associations between genetically proxied inhibition of six putative ibrutinib drug targets (ErbB2/HER2, CSK, JAK3, TEC, BLK, and PLCG2) and the atrial fibrillation (AF) risk, proarrhythmic ECG indices, and cardiometabolic traits and diseases. Inverse-variance weighted random-effects models and Wald ratio were used to examine the associations between genetically proxied inhibition of these drug targets and the risk of outcomes. Colocalization analyses were employed to examine the robustness of the causally significant findings. ELISAs were used to measure ErbB2 levels in intracardiac plasma samples.
Results
Genetically proxied ErbB2 inhibition was associated with an increased AF risk, higher P wave terminal force, and prolonged QTc interval. Patients with AF had significantly higher intracardiac ErbB2 levels compared with patients with paroxysmal supraventricular tachycardia. CSK inhibition prolonged the QRS duration, decreased the QTc interval, and was potentially linked to conduction blocks. PLCG2 inhibition led to decreased P wave terminal force, shorter QTc interval, and increased risk of left bundle branch block. BLK inhibition shortened the QTc interval and was also associated with atrioventricular block.
Conclusion
The off-target effects and downstream targets of ibrutinib, including CSK, PLCG2, ERBB2, TEC, and BLK, may lead to cardiac electrical homeostasis imbalances and lethal cardiovascular diseases. Using drugs that inhibit these targets should be given extra caution.
Abbreviations
-
- AF
-
- atrial fibrillation
-
- BLK
-
- B-lymphoid kinase
-
- BMI
-
- body mass index
-
- BTK
-
- Bruton's tyrosine kinase
-
- CSK
-
- C-terminal src kinase
-
- DBP
-
- diastolic blood pressure
-
- ErbB2
-
- human epidermal growth factor receptor 2
-
- GWAS
-
- genome-wide association study
-
- HDL-C
-
- high-density lipoprotein cholesterol
-
- JAK3
-
- Janus kinase 3
-
- LBBB
-
- left bundle branch block
-
- LDL-C
-
- low-density lipoprotein cholesterol
-
- PLCG2
-
- phospholipase C γ 2
-
- PSVT
-
- paroxysmal supraventricular tachycardia
-
- QTL
-
- quantitative trait loci
-
- RBBB
-
- right bundle branch block
-
- SBP
-
- systolic blood pressure
-
- TEC
-
- T-cell kinase
1 Introduction
Atrial fibrillation (AF) is the most common sustained arrhythmia that impacts many patients worldwide. AF is characterized by nonchronological excitation of the atria, leading to dyssynchronous atrial excitation and ventricular contraction. Although community-based cohort studies have provided critical insights into the associated risk factors [1], AF can also be iatrogenic.
Bruton's tyrosine kinase inhibitor (BTKi) ibrutinib is a first-line drug prescribed to treat B-cell neoplasms but has also been shown to increase the risks of AF [2], hypertension [3], and bleeding [4]. Real-world clinical trials have suggested that the rate of ibrutinib-induced AF is as high as 16% to 25% [5, 6]. Notably, AF inducibility was significantly lower when using acalabrutinib [7, 8], a more selective BTKi than ibrutinib, suggesting that BTK-independent pathways may mediate AF susceptibility. In addition to BTK, ibrutinib can covalently bind in varying degrees to the C481 thiol group of kinases [9, 10]. This includes the T-cell kinase (TEC) kinase family, EGFR kinase family, tyrosine kinases, and SRC kinase family.
It is not well understood if inhibiting these kinases can lead to increased AF. Therefore, we used the Mendelian randomization (MR) approach to examine the effects of long-term inhibition of the human epidermal growth factor receptor 2 (ErbB2/HER2), C-terminal src kinase (CSK), Janus kinase 3 (JAK3), TEC, B-lymphoid kinase (BLK), and BTK downstream target phospholipase C γ 2 (PLCG2) on AF risk, conduction blocks, proarrhythmic electrocardiogram indices, and cardiometabolic traits/diseases. Because the gene encoding BTK is on the X chromosome, data are not available in most exposure and outcome datasets.
2 Methods
2.1 Study Population
2.1.1 AF and Electrocardiogram Indices
Summary genetic association data of AF were obtained from a meta-analysis of genome-wide association study (GWAS) results from the Nord-Trøndelag Health Study (HUNT), deCODE, MGI, DiscovEHR, UK Biobank, and AFGen Consortium [11]. This meta-analysis resulted in up to 60,620 European ancestry cases and 970,216 European ancestry controls. Associations with AF in these cohorts were adjusted for age, sex, principal components, and genotyping batch.
For replication analyses, summary genetic association data were obtained for 22,068 AF cases and 116,926 controls of European ancestry from the FinnGen cohort [12]. We also examined if the findings could be extended to individuals of East Asian ancestry by obtaining summary genetic association data of 8180 AF cases and 28,612 controls of Japanese ancestry from Biobank Japan [13].
The 12-lead electrocardiogram indices were taken at rest in the supine position, including P wave duration [14] (n = 37,678), P wave terminal force [14] (n = 33,955), PR interval [15] (n = 271,570), QRS duration (n = 10,815, http://www.nealelab.is/uk-biobank/), QTc [16] (n = 84,630), and heart rate variability (standard deviation of all normal RR [NN] intervals during a 24-h period [SDNN] n = 27,850, root mean square of successive differences between normal heartbeats [RMSSD] n = 26,523, and the peak-valley respiratory sinus arrhythmia or high-frequency power [pvRSA/HF] n = 24,088) [17]. All algorithms that were used to generate these indices were described in detail in the respective papers.
2.1.2 Conduction Blocks
Atrioventricular block (n = 2388/156,711), left bundle branch block (LBBB, n = 776/156,711), and right bundle branch block (RBBB, n = 405/156,711) were obtained from the FinnGen cohort [12].
2.1.3 Cardiometabolic Traits and Cardiovascular Diseases
In addition, 17 cardiometabolic factors were studied, including lipids (total cholesterol, triglycerides, high-density lipoprotein cholesterol [HDL-C], low-density lipoprotein cholesterol [LDL-C], apolipoprotein A1 and B, and lipoprotein [a]) [18, 19], blood pressure (systolic blood pressure [SBP] and diastolic blood pressure [DBP]) [20], glycemic traits (fasting glucose, fasting insulin, 2-h post-challenge glucose, and HbA1c) [21, 22], and anthropometric traits (body mass index [BMI], waist circumference, hip circumference, and waist-to-hip ratio) [23]. Additionally, 11 cardiometabolic diseases were included in the analysis, including stroke and stroke subtypes (large artery atherosclerosis, small-vessel, and cardioembolic) [24], coronary artery disease [25], heart failure [26], aortic aneurysms [12], chronic kidney disease [27], type 1 diabetes [28], and type 2 diabetes [29]. Most participants in these cohorts were of European ancestry (Table S1).
2.2 Instrument Construction
To proxy ErbB2, CSK, JAK3, TEC, BLK, and PLCG2 inhibition, summary cis-expression quantitative trait loci (cis-eQTL) data were obtained from the eQTLGen Consortium, which was generated from peripheral blood samples of 31,684 individuals of European ancestry. We also obtained tissue-specific cis-eQTL data from the Genotype-Tissue Expression project [30] (GTEx, v8 release) and cis-pQTL data from deCODE genetics [31] (only data for CSK and PLCG2 were available) to replicate the analyses. Genome-wide significant single nucleotide polymorphisms (SNPs) (p < 5 × 10−8) in weak linkage disequilibrium (r2 < 0.001, based on the 1000 Genomes Phase 3 European reference panel) within ±100 kb from the target gene loci that were associated with the gene expression were obtained. In addition, we obtained genome-wide significant variants that were associated with blood pressure in previously described GWAS analyses and unadjusted UK Biobank GWAS analyses (without adjustments for BMI or antihypertensive medication use) to explore the CSK proxied blood pressure effect on AF. The SNP used for this was also examined to determine if it is an eQTL for the gene. F-statistic < 10 was used to exclude SNPs as weak instruments, with results that are likely to be influenced by weak instrument bias because of a limited proportion of the variance explained in a drug target (Table S2).
2.3 MR
Variant–exposure association data were individually harmonized with variant–outcome association using the TwoSampleMR package. During harmonization, the positive strand allele was inferred using allele frequencies. Only SNPs presented in both variant–exposure and variant–outcome associations were used in the analysis. This study is reported as per the Guidelines for strengthening the reporting of MR studies (STROBE-MR) checklist (Additional file 1 STROBE Checklist) [32].
2.4 Statistical Analyses
All statistical analyses were performed in R version 4.2.3, including the “TwoSampleMR (version 0.6.3)” and “coloc (version 5.2.3)” packages. Wald ratio estimates were employed to estimate the causal effects of genetically proxied drug target inhibition when only one SNP was used as an instrument variant on outcomes. Inverse-variance weighted (IVW) random-effects models were used when there was more than one SNP.
Colocalization was performed to examine if the drug targets and outcomes showing significant causality in MR analyses share the same causal variant at a given locus. Colocalization analysis was performed with ±100 kb windows from the gene locus to proxy the respective drug targets. A posterior probability of > 0.95 was considered significant. Summary-data-based MR (SMR) and HEIDI tests [33] were used to further validate the pleiotropic association between gene expression and outcomes.
To avoid false positives resulting from multiple comparisons, a Bonferroni correction was used to establish a p threshold of 2.14 × 10−4 (false positive rate = 0.05/234, 6 drug targets tested against 39 outcomes). A p < 2.14 × 10−4 was defined as a “robust association,” with 2.14 × 10−4 < p < 0.05 defined as a “potential association.” For cardiovascular outcomes, the results are presented as odds ratios (ORs) with respective 95% confidence intervals (CIs). Continuous cardiometabolic measures are presented as coefficients (β) and 95% CI.
2.5 Intracardiac ERBB2/HER2 Quantification
This study included a total of 34 patients with paroxysmal AF (PaAF, n = 18) and persistent AF (PeAF, n = 16) who underwent their first radiofrequency ablation procedure in the Department of Cardiology of Peking University First Hospital from January 2021 to July 2022. As a control group, patients with left-sided accessory pathways combined with paroxysmal supraventricular tachycardia (PSVT, n = 11) were also included. Left atrial whole blood (10 mL) was collected and added into an EDTA anticoagulation tube, after which 10 μL of protease inhibitor was immediately added. The specimen was then shaken gently and inverted several times for homogenization. After centrifugation at 3000 r/min for 15 min at 4°C, the upper plasma layer was collected and dispensed into microcentrifuge tubes, of which 1 mL was centrifuged a second time. The supernatant was removed by centrifugation at 12,000 × g for 10 min at 4°C and transferred to a new LoBind tube. All specimens were then stored at −80°C. The human ErbB2/HER2 DuoSet ELISA kit (Bio-Techne/R&D Systems, Minneapolis, Minnesota, the United States, DY1129B) was used to quantify ErbB2/HER2 levels following the manufacturer's suggested protocol.
3 Results
3.1 AF and Cardiometabolic Outcomes
The characteristics and F-statistics of the six drug targets were examined for their respective SNPs ranging from 35 to 1431, suggesting that our analyses were unlikely to suffer from weak instrument bias. The r2 and F-statistic estimates for the instrument variables of each target are presented in Tables S2–S4.
Breast cancer was used as a positive control for ErbB2 (all positive controls are presented in Table S5), with rs903506 found to be associated with a lower risk of HER2-positive breast cancer (FinnGen, n = 4263/99,267, OR: 0.51, 95% CI: 0.32–0.83, p = 0.006). Genetically proxied ErbB2 inhibition was associated with an increased risk of AF (Wald ratio, per fold change expression: OR: 1.49, 95% CI: 1.30–1.71, p = 8.4 × 10−9) and lower levels of HDL-C (β = −0.23, 95% CI: −0.32–−0.13, p = 7.1 × 10−6), total cholesterol (β = −0.21, 95% CI: −0.31–−0.11, p = 3.0 × 10−5), and apolipoprotein A1 (β = −0.17, 95% CI: −0.24–−0.09, p = 2.7 × 10−5) (Figure 1). In addition, left atrial serum ErbB2 levels were significantly higher in patients with AF than in patients with PSVT (467.27 ± 118.97 pg/mL vs. 577.29 ± 157.18 pg/mL, p = 0.04, participant characteristics shown in Table S6).
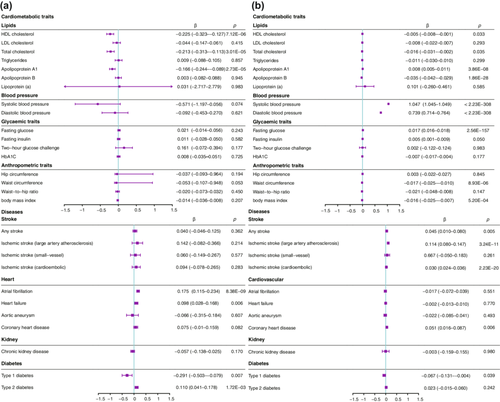
Genetically proxied CSK inhibition was associated with higher SBP (IVW, β = 1.05, 95% CI: 1.05–1.05, p < 2.2 × 10−308) and DBP (β = 0.74, 95% CI: 0.71–0.76, p < 2.2 × 10−308), with less robust effects on apolipoprotein A1 and apolipoprotein B (Figure 2). However, genetically proxied CSK inhibition was not associated with the risk of AF (OR: 0.98, 95% CI: 0.93–1.04; p = 0.55). Because genetically proxied CSK inhibition was associated with an increased risk of cardioembolic ischemic stroke (OR: 1.03, 95% CI: 1.02–1.04, p = 2.2 × 10−20), CSK inhibition proxied SBP (rs112297944) was used to explore the association with AF (Table S7). However, CSK inhibition proxied SBP was not associated with AF (OR equivalent to 1 mmHg higher SBP: 0.99, 95% CI: 0.95–1.02, p = 0.50), which was consistent with cis-eQTL proxied CSK inhibition. Using SNPs from the unadjusted SBP GWAS data from the UK Biobank showed similar results (OR equivalent to 1 mmHg higher SBP: 2.73, 95% CI: 0.78–5.81, p = 0.14).
Genetically proxied inhibition of JAK3, BLK, TEC, and PLCG2 were not associated with AF (Table S8). Genetically proxied BLK inhibition was associated with higher fasting glucose and total cholesterol levels, which are risk factors for AF.
3.2 Proarrhythmic Electrocardiogram Indices and Conduction Blocks
All targets except JAK3 were significantly associated with various proarrhythmic electrocardiogram indices (Tables 1 and 2). Genetically proxied BLK, PLCG2, and CSK inhibition were significantly associated with higher atrioventricular block, higher LBBB, and lower RBBB, respectively. Interestingly, genetically proxied CSK inhibition was associated with lower RBBB rather than higher LBBB (Table 3).
| ECG traits | eQTL source | No. of SNPs | Methods | β (95% CI) | p |
|---|---|---|---|---|---|
| P wave duration (ms) | |||||
| PLCG2 | Blood | 1 | Wald ratio | 0.75 (0.13–1.37) | 0.02 |
| P wave terminal force (μV × ms) | |||||
| ErbB2 | Atrial appendage | 1 | Wald ratio | 174.17 (8.34–339.99) | 0.03 |
| PLCG2 | Blood | 1 | Wald ratio | −347.69 (−698.96–−5.41) | 0.05 |
| TEC | Blood | 1 | Wald ratio | 592.24 (61.28–1123.21) | 0.03 |
| PR duration (ms) | |||||
| PLCG2 | Blood | 3 | IVW | 0.16 (0.06–0.26) | 0.002 |
| QRS duration (ms) | |||||
| CSK | Blood | 2 | IVW | 0.04 (0.03–0.06) | 2.10 × 10−11 |
| ErbB2 | Blood | 1 | Wald ratio | −0.29 (−0.55– −0.03) | 0.03 |
| QTc interval (ms) | |||||
| ErbB2 | Atrial appendage | 1 | Wald ratio | 2.61 (1.17–4.05) | 3.67 × 10−4 |
| Blood | 1 | Wald ratio | 4.22 (1.90–6.55) | 3.71 × 10−4 | |
| PLCG2 | Atrial appendage | 1 | Wald ratio | −1.08 (−1.88– −0.29) | 0.01 |
| Blood | 3 | IVW | −0.79 (−1.26– −0.32) | 0.001 | |
| CSK | Blood | 2 | IVW | −0.78 (−1.25– −0.30) | 0.001 |
| BLK | Blood | 2 | IVW | −0.21 (−0.23– −0.19) | 1.61 × 10−80 |
- Abbreviations: BLK, B-lymphoid kinase; CSK, C-terminal src kinase; ErbB2, human epidermal growth factor receptor 2; IVW, inverse-variance weighted model; JAK3, Janus kinase 3; PLCG2, phospholipase C γ 2; TEC, T-cell kinase.
| Heart rate variability | No. of SNPs | Methods | Odd ratio (95% CI) | p |
|---|---|---|---|---|
| RMSSD | ||||
| CSK | 2 | IVW | −0.01 (−0.03–−0.003) | 0.04 |
| PLCG2 | 3 | IVW | −0.04 (−0.05–−0.02) | 2.01 × 10−4 |
- Abbreviations: CSK, C-terminal src kinase; IVW, inverse-variance weighted model; PLCG2, phospholipase C γ 2; RMSSD, root mean square of successive differences.
| Conduction blocks | No. of SNPs | Methods | Odd ratio (95% CI) | p |
|---|---|---|---|---|
| Atrioventricular block | ||||
| BLK | 2 | IVW | 1.11 (1.11–1.11) | < 2.23 × 10−308 |
| Left bundle branch block | ||||
| CSK | 2 | IVW | 1.22 (1.21–1.23) | < 2.23 × 10−308 |
| PLCG2 | 3 | IVW | 0.76 (0.63–0.93) | 0.006 |
| Right bundle branch block | ||||
| CSK | 2 | IVW | 0.91 (0.89–0.93) | 1.66 × 10−15 |
- Abbreviations: BLK, B-lymphoid kinase; CSK, C-terminal src kinase; IVW, inverse-variance weighted model; PLCG2, phospholipase C γ 2.
3.3 Sensitivity Analysis
The FinnGen cohort of AF and flutter (Ncase/Ncontrol = 22,068/116,926), FinnGen cohort of AF and flutter reimbursement (Ncase/Ncontrol = 22,068/116,926), and Biobank Japan cohort of AF (Ncase/Ncontrol = 8180/28,612) were used to replicate the association of ErbB2 and AF. ErbB2 expression was potentially associated with increased risk of AF and flutter (OR: 1.24, 95% CI: 1.02–1.50, p = 0.03) and AF reimbursement (OR: 1.34, 95% CI: 1.04–1.71, p = 0.02) in the FinnGen cohort. These results were not replicated in the Japanese population (OR: 1.15, 95% CI: 0.90–1.47, p = 0.25).
In addition, cis-eQTL ErbB2 (rs2941504) from the atrial appendage replicated the association of ErbB2 with AF (Table S9). The MR associations between the cis-pQTL of CSK/PLCG2 and outcomes are presented in Table S10. PLCG2 inhibition was associated with a higher AF risk, LBBB, and longer QTc interval. The CSK inhibition results with cis-pQTL were similar to those with cis-eQTL.
3.4 Colocalization and SMR
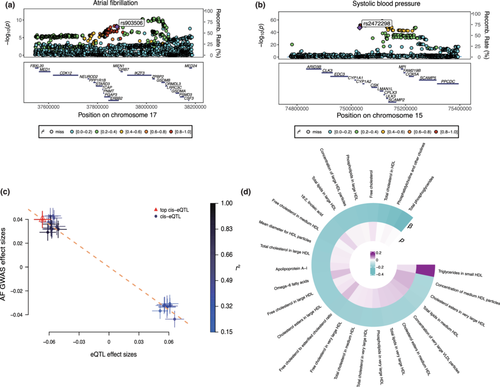
Colocalization analysis suggested that ErbB2 expression and AF associations had a 100% posterior probability of sharing a causal variant within the ErbB2 locus (Table S11). Regional Manhattan plots examining all SNPs ± 300 kb from the top SNP for ErbB2 expression (rs903506) for their associations with AF (Figure 2a) did not appear to support the presence of two or more independent causal variants driving the associations across either trait. Colocalization analysis suggested that CSK expression had a 99.9% posterior probability of sharing a causal variant with SBP, QRS duration, LBBB, and RBBB within the CSK locus (Table S11). BLK expression had a 99.9% posterior probability of sharing a causal variant with the atrioventricular block within the BLK locus. SMR analysis showed that genetically determined expression of ErbB2 was associated with AF (βSMR = −0.60, pSMR = 2.6 × 10−6, and pHEIDI = 0.69, Figure 2c,d).
4 Discussion
In this study, we aimed to use MR to strengthen our understanding of the effects of putative ibrutinib targets on arrhythmia risk and pro-arrhythmia risk factors. We observed that AF risk was associated with ErbB2 and potentially PLCG2. The intracardiac serum ErbB2 levels were significantly higher in patients with AF than in patients with PSVT. In addition, CSK, PLCG2, and BLK were robustly associated with proarrhythmic ECG indices, conduction blocks, and cardiometabolic traits, which have not been previously reported. These results support a causal nature of the associations between ibrutinib and a variety of arrhythmias/proarrhythmic traits and indices. Furthermore, the proarrhythmic impact of ibrutinib may be from a combination of off-target effects and downstream pathway inhibition. Inhibition of these targets may contribute to cardiac electrical homeostasis imbalances, potentially explaining why ibrutinib is more proarrhythmic than acalabrutinib.
MR infers causal effects of exposures if the critical assumptions related to instrument validity are met. Our results were consistent across exposures derived from blood and tissue at the RNA and protein levels, as well as with outcomes from different cohorts of European descendants. This indicated a robust association between the exposures and outcomes. In addition, only cis variants with low linkage disequilibrium were included to avoid horizontal pleiotropy, while colocalization provided further strong evidence.
Ibrutinib, a potent BTK inhibitor, exhibits irreversible binding to BTK, thereby impeding downstream signaling that results in reduced cell proliferation and survival rates of B-cell-related malignancies. As a first-generation BTK inhibitor, ibrutinib has demonstrated notable clinical efficacy across various B-cell malignancies. While generally well-tolerated with predominantly mild and manageable side effects, ibrutinib is associated with serious adverse effects, notably AF [34]. According to previous research by Ganatra et al. [35], 190 of 2166 (8.15% adjusted incidence weighted according to sample size) patients receiving ibrutinib developed AF over a median follow-up period of 18.32 months. The incidence of ibrutinib-associated AF was 5.77 per 100 person-years, which is significantly higher compared with the general population risk (AF incidence of approximately 1.5%–2%). Moreover, there were no previous clinical reports describing an ibrutinib-induced conduction delay. Tuomi et al. [36] showed that ibrutinib reduced the action potential upstroke velocity and sodium current in isolated mice atrial myocytes. This was not observed in acalabrutinib-treated atrial myocytes, which could potentially account for the conduction delay induced by ibrutinib. The precise mechanisms governing ibrutinib-induced AF and arrhythmias remain elusive, but it is commonly postulated that the inhibition of cardiac off-target molecules, including ErbB2, CSK-SFKs, and BLK signaling, contributes to its proarrhythmic effects [37, 38].
ErbB2, also known as HER2, is required in atrial electrical conduction during embryonic development [39]. Additionally, activating neuregulin/ErbB2/ErbB4 signaling may induce cardiomyocyte proliferation and heart injury repair [40, 41]. However, no experimental study to our knowledge has explored the proarrhythmic effect on ErbB2. A previous study [42] showed that ErbB2 mRNA expression levels are upregulated in the left atrial appendage, which is consistent with our intracardiac serum ErbB2 quantification data. Despite the absence of pQTL data for ErbB2, our findings using the eQTL data from blood and atrial appendage showed that ErbB2 inhibition increased the P wave terminal force, QTc interval, and AF risk. However, the AF incidence in breast cancer patients receiving trastuzumab (a monoclonal antibody targeting Her2/ErbB2) was far less than that in patients with hematologic neoplasms receiving ibrutinib [43]. In contrast, heart failure and loss of left ventricular contractile function were more common in patients treated with trastuzumab [44], which is consistent with our results (Table S8). Henceforth, it appears unlikely that exclusive inhibition of ErbB2 is the singular etiological factor contributing to ibrutinib-induced arrhythmia.
Cardiac-specific CSK knockout has been previously reported to be the mechanism through which ibrutinib leads to AF [45]. Although our data did not yield a direct causal association with AF, we found that CSK inhibition prolonged the QRS duration, decreased the QTc interval, and was potentially linked to conduction blocks. This may have been from its effects on potassium homeostasis [46]. In addition, CSK deficiency has been shown to trigger augmented aldosterone production, which in turn increases sodium reabsorption and leads to hypertension [47]. These results were replicated in our study, but a causal relationship with AF was not established using the hypertensive effect proxied by CSK inhibition.
PLCG2 is a kinase that acts downstream of BTK in the B-cell receptor signaling pathway. Only a few genetic studies [48, 49] have discussed the relationship between PLCG2 and arrhythmias. While a causal relationship with AF was not validated using pQTL and eQTL as instrumental variables in this study, consistent findings emerged with the increased P wave terminal force, shorter QTc interval, and increased risk of LBBB. Despite the established association of PLCG2 with inflammation and immunity [50], the biological mechanisms through which it may induce arrhythmias remain unclear.
BLK is a member of the SRC family protein tyrosine kinases (SFKs), a subfamily of non-receptor tyrosine kinases. Nilotinib and dasatinib, among other SFK inhibitors, have been associated with QTc abnormalities [51] and atrioventricular block [52]. However, studies have also shown that c-SRC inhibition could improve connexin 43 levels and conduction velocity [53]. Our findings were consistent with clinical research on ibrutinib, indicating an association between BLK, CSK, PLCG2, and shorter QTc intervals [52].
Overall, our results indicated widespread correlations between these targets and cardiometabolic traits, such as HDL-C, total cholesterol, and fasting blood glucose levels. Changes in these traits pose risks not only for arrhythmias but also for other lethal cardiovascular diseases. This results in a significant threat to the well-being of patients using drugs that target these specific proteins.
Although MR is a powerful tool for inferring causality, it has several inherent limitations. Even though colocalization analysis and the HEIDI test were used to minimize possible bias caused by linkage disequilibrium, horizontal pleiotropy could still affect our results. This would violate the core assumptions of MR because we were unable to identify the eQTL association for all phenotypes. Restricting the study population to European ancestry minimized the population structure bias, but it also limited the generalization of our findings to other populations. We employed only six putative targets, with ibrutinib displaying varying inhibition capacity and affinity toward these targets. MR is a genetic proxy for lifelong inhibition and cannot capture the impact of drug use in a clinical setting, resulting in potential exaggerations of the actual effects of ibrutinib on arrhythmias. While our study had some other limitations, such as a lack of protein-level instrumental variables, the absence of replicated studies for electrocardiographic outcomes and conduction blocks, and a limited number of SNPs for each target, our results underwent rigorous p corrections and validations from multiple exposure sources. Therefore, the conclusions drawn should be considered relatively robust.
5 Conclusion
In summary, the off-target effects and downstream targets of ibrutinib, including CSK, PLCG2, ERBB2, TEC, and BLK, may lead to arrhythmias and other serious cardiovascular diseases. Using drugs that inhibit these targets should be approached with extra caution.
Author Contributions
Hongxuan Xu: conceptualization (lead), data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), writing–original draft (lead), writing–review and editing (equal). Bingxun Li: conceptualization (supporting), formal analysis (equal), investigation (equal), writing–review and editing (equal). Pinchao Lv: data curation (equal), investigation (equal), project administration (equal). Ying Chen: project administration (equal). Yanyun Lin: project administration (supporting). An Zhang: project administration (supporting). Jing Zhao: investigation (supporting), project administration (supporting). Guoxiong Zhou: investigation (supporting). Lin Wu: funding acquisition (lead), supervision (lead), validation (equal).
Acknowledgments
The authors would like to thank all the GWAS participants included in this study.
Ethics Statement
All contributing GWAS studies received the approval of the respective institutional review board in accordance with the Declaration of Helsinki. The use of human samples in this study was approved by the Institutional Review Board of the Peking University First Hospital (Beijing, China; Nos. 2020-360 and 2020-451) and was conducted following the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice.
Consent
All participants provided informed consent.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Additional information on statistical analysis, imputation, and quality control measures are included in the respective publications. All data used in this study can be downloaded from the cited publications. GWAS summary data from UK Biobank can be accessed at: https://www.nealelab.is/uk-biobank. FinnGen: https://www.finngen.fi/en/access_results. Gtex eQTL: https://www.gtexportal.org/home/downloads/adult-gtex#qtl. Decode pQTL: https://www.decode.com/summarydata/. QTc interval: https://personal.broadinstitute.org/ryank/Nauffal_2022_QT_GWAS_SAIGE.zip. Summary GWAS data of P wave duration, P wave terminal force, PR interval, heart rate variability, and atrial fibrillation can be accessed at GWAS catalog under the ID (GCST004826, GCST004824, GCST010320, GCST004732, GCST004733, GCST004734, and GCST006414).




