The Utility of Long-Read Sequencing in Diagnosing Early Onset Parkinson's Disease
Abstract
Objective
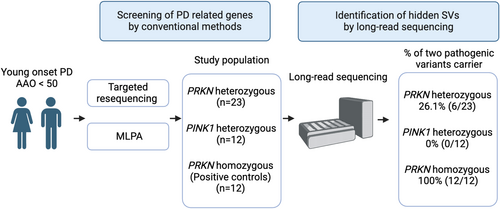
Variants in PRKN and PINK1 are the leading cause of early-onset autosomal recessive Parkinson's disease, yet many cases remain genetically unresolved. We previously identified a 7 megabases complex structural variant in a pair of monozygotic twins using Oxford Nanopore Technologies (ONT) long-read sequencing. This study aims to determine if ONT long-read sequencing can detect a second variant in other unresolved early-onset Parkinson's disease (EOPD) cases with 1 heterozygous PRKN or PINK1 variant.
Methods
ONT long-read sequencing was performed on EOPD patients with 1 reported PRKN/PINK1 pathogenic variant, with onset age under 50. Positive controls included EOPD patients with 2 known PRKN pathogenic variants. Initial testing involved short-read targeted panel sequencing for single nucleotide variants and multiplex ligation-dependent probe amplification for copy number variants.
Results
A total of 47 patients were studied (PRKN “one-variant,” n = 23; PINK1 “one-variant,” n = 12; PRKN “two-variants,” n = 12). ONT long-read sequencing identified a second pathogenic variant in 26% of PRKN “one-variant” patients (6/23), but none in PINK1 “one-variant” patients (0/12). Detected variants included 1 complex inversion, 2 structural variant overlaps, and 3 duplications. In the PRKN “two-variants” group, both variants were identified in all patients (100%, 12/12).
Interpretation
ONT long-read sequencing effectively identifies pathogenic structural variants in the PRKN locus missed by conventional methods. It should be considered for unresolved EOPD cases when a second variant is not detected through conventional approaches. ANN NEUROL 2025;97:753–765
Parkinson's disease (PD) is a neurodegenerative disorder showing motor symptoms, including resting tremor, rigidity, bradykinesia, and postural instability. These motor symptoms are caused by loss of dopaminergic neurons in the substantia nigra pars compacta. PD is considered to be caused by a combination of genetics, environment, and aging.1
Approximately 5 to 10% of all PD cases can be attributed to a “monogenic” cause of disease.2 Biallelic PRKN and PINK1 mutations are known to be the frequent cause of early onset PD (EOPD) and autosomal recessive PD.3, 4 The frequency of PRKN mutations increases with lower age at onset (AAO) of PD and is estimated to account for 77% in the PD patients with AAO younger than 20.5 Typically, biallelic PRKN and PINK1 variant carriers are characterized by early onset parkinsonism, foot dystonia, sleep benefit, and good response to levodopa.6, 7
Intriguingly, monoallelic variants (carriers of 1 damaging variant) of PRKN and PINK1 have been considered to be associated with PD.8-11 Monoallelic PRKN or PINK1 carriers are estimated to account for 2% of all PD patients.12 However, some studies showed a negative association with PD and heterozygous variants of PRKN and PINK1.13-15 Therefore, the role of monoallelic PRKN/PINK1 variants remains controversial.
PRKN and PINK1 can harbor pathogenic variants, including single nucleotide variants (SNVs), exon dosage variations, and complex rearrangements.16 Using long-read sequencing, we recently identified a heterozygous large inversion of PRKN, which was missed by multiplex ligation-dependent probe amplification (MLPA) and short-read targeted resequencing in monozygotic twins with EOPD known to have a heterozygous exon 3 deletion.17 Expanding on this, using long-read sequencing, we assessed how often complex structural variants (SVs), like inversions, are missed by short-read sequencing and MLPA in young-onset PD patients who only carry monoallelic PRKN and PINK1 variants.
Methods
Study Design and Participants
All the participants were selected according to the following criteria: (1) AAO of PD is younger than the age of 50, (2) the participant was confirmed to have 1 pathogenic variant in PRKN or PINK1 based on the targeted resequencing of PD-related genes and MLPA, (3) the participant does not have any pathogenic variants in other known PD or dementia-related genes (SNCA, UCHL1, PARK7, LRRK2, ATP13A2, GIGYF2, HTRA2, PLA2G6, FBXO7, VPS35, EIF4G1, DNAJC6, SYNJ1, DNAJC13, CHCHD2, GCH1, NR4A2, VPS13C, RAB7L1, BST1, C19orf12, RAB39B, MAPT, PSEN1, GRN, APP, and APOE). We also included patients with 2 known PRKN variants to assess the overall performance of long-read sequencing to detect these variants.
All the participants underwent a neurological examination and clinical information was collected by the attending neurologist. PD was clinically diagnosed according to standard clinical criteria.18, 19 DNA was extracted from peripheral blood using the standard protocol using QIAamp DNA Blood Maxi Kit (QIAGEN, Venlo, The Netherlands). The study design is visualized in Figure 1.
The study was approved by the ethics committee of Juntendo University, Tokyo, Japan, and all participants provided written informed consent to participate in the research described in this study (M08-0477-M09).
Genetic Testing
Targeted Panel Sequencing Using Short-Read Sequencing
The targeted panel sequencing was performed to sequence for PD-related genes have been previously reported.11 In brief, the Ion Torrent system (Thermo Fisher Scientific, Waltham, MA) was used for sequencing, and we selected rare variants with an allelic frequency under 0.001 for autosomal dominant inheritance and under 0.005 for autosomal recessive inheritance by referring to public gene databases and annotating them using several prediction tools to define the pathogenicity of the variant.
SVs Screening
Initially, copy number variants (CNVs) of PRKN were analyzed by quantitative polymerase chain reaction (qPCR) with TaqMan probe (Applied Biosystems, Foster City, CA) using ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA) or by multiplex MLPA using SALSA MLPA Probemix P051 Parkinson mix (MRC-Holland, Amsterdam, The Netherlands), as we reported previously.11, 20 Second, for cases where we found either 1 variant of PRKN or PINK1 after targeted resequencing and the initial qPCR/MLPA analysis, we conducted a second MLPA experiment. This involved using the MLPA P052 Parkinson probe mix (MRC-Holland, Amsterdam, The Netherlands) to standardize the method for screening CNVs. We determined the number of PRKN/PINK1 CNVs by considering the results from both the initial (qPCR/MLPA with P051) and the second (MLPA with P052) screenings. MLPA procedures were performed according to the manufacturer's instructions.
Oxford Nanopore Technologies Long-Read Sequencing
We used the DNA prepared for the short-read sequencing for the long-read sequencing. Sequencing was prepared according to our protocol reported previously.21, 22 In brief, DNA samples were sized using the Femto Pulse (Agilent Technologies Santa Clara, CA) and run on the Sage BluePippin system (Sage Science, Beverly, MA) to remove DNA fragments below 10 kb. Libraries were prepared using the Kit V14 Ligation sequencing kit from Oxford Nanopore Technologies (ONT) and sequenced using PromethION for 72 hours on an R10.4.1 flow cell (Oxford Nanopore Technologies, Oxford, UK). Base calling was performed by Dorado v0.3.4 (https://github.com/nanoporetech/dorado), and Minimap v2.26 was used to map the reads to the GRCh38 reference genome. Sniffles v2.2 and CuteSV v2.0.3, and Seversus v0.1.2 were used for calling SVs.23-25 SVs were annotated by AnnotSV v3.1.1.26 All the identified variants in at least 1 SV caller were confirmed visually by Integrative Genome Viewer (IGV).27 SNVs were called by Clair3, and the output vcf was annotated using Annovar.28, 29 To phase the variants, PEPPER-Margin-Deep-Variant v0.8 was used with -phased_output option.30 Additional adaptive long-read sequencing for PRKN and PINK1 was performed in 12 samples to increase the coverage of PRKN and PINK1 using ONT-recommended guidelines.
Confirmation of SVs
To confirm the complex SVs that were only identified by long-read sequencing, we amplified the breakpoints region using PCR and performed Sanger sequencing by the primers specifically designed by Primer 3 (Table S1).
Statistics
We compared the clinical phenotype between PRKN one-variant carriers and PRKN two-variants carriers after ONT long-read sequencing by Pearson's correlation coefficient and point-biserial correlation coefficient.
Results
Data Overview
We included 47 patient samples (PRKN one-variant group n = 23, PRKN two-variants group n = 12, PINK1 one-variant group n = 12; female:male = 28:20) in this study (Table 1). All the samples satisfied the DNA quality criteria for inclusion (Tables S2–S4). The overall data output for long-read sequencing is 98.5 ± 25.04 Gb, and N50 was 20.3 ± 2.82 kb for genome sequencing, and for adaptive sampling, the data output is 13.1 ± 7.17 Gb, and N50 was 1.60 ± 1.68 kb (Tables S5 and S6, Fig S1).
| PRKN hetero | PINK1 hetero | PRKN homo | |
|---|---|---|---|
| One-variant group | One-variant group | Two-variants group | |
| No. | 23 | 12 | 12 |
| Age at onset | 37.2 ± 7.99 | 32.7 ± 9.70 | 27.7 ± 10.06 |
| Age at examination | 50.3 ± 12.40 | 46.3 ± 13.55 | 47.4 ± 13.68 |
| Female:male | 15:8 | 5:7 | 7:5 |
| Known heterozygous variant (SNV/SV) | 9/14 | 12/0 | 4/20 |
- hetero = heterozygous; homo = homozygous; SNV = single nucleotide variant; SV = structural variant.
Assessing the Performance of Long-Read Sequencing in PRKN-PD
To assess the overall performance of long-read sequencing, we included 12 PRKN patient samples carrying 2 known PRKN variants. All the known variants, including SNVs and SVs identified by panel sequencing, qPCR, and MLPA were identified and confirmed using long-read sequencing (Table 2), showing 100% accuracy. In 1 case (PRKN-18), in which duplication of exon 2 and deletion of exon 5 were found by MLPA, long-read sequencing was able to define the genomic events more accurately and reported that there was a duplication of exons 2 to 4 and deletion of exon 3 to 5. In 3 cases (PRKN-12, PRKN-23, and PRKN-26), SVs were identified as duplications by MLPA but were categorized as inversions by long-read sequencing SV callers. Visual inspection using the IGV indicated that 1 allele of the duplication was inverted, similarly showing the value of MLPA but also the superiority of long-read sequencing in defining what the actual variant is. Additionally, all known SVs and SNVs from the PRKN one-variant group were identified by long-read sequencing. We screened monogenic PD-related genes other than PRKN and PINK1 in all samples and found no pathogenic variant.
| One variant of PRKN in MLPA and targeted resequencing | Two variants of PRKN in MLPA and targeted resequencing | One variant of PINK1 in MLPA and targeted resequencing | |
|---|---|---|---|
| No. | 23 | 12 | 12 |
| Two variants carriers by long-read sequencing | 26.1% (6/23) | 100% (12 /12) | 0% (0/12) |
| Variants LRS missed | 0 | 0 | 0 |
| Discordant with LRS and MLPA | 8a | 1b | 0 |
- a Including 6 newly identified two-variant carriers, 1 with complex variant (DUP-NML-DUP/INV), and 1 with size difference of deletion between long-read sequencing and MLPA.
- b One sample has an overlapping copy number variant on different alleles, which makes the size of structural variants different between long-read sequencing and MLPA.
- DUP = duplication; INV = inversion; LRS = long-read sequencing; MLPA = multiplex ligation-dependent probe amplification; NML = normal.
Identification of the Second PRKN Variant in Patients with a Single PRKN Variant
In the other 35 patients with 1 PRKN or PINK1 variant, long-read sequencing identified a second, previously undetected pathogenic variant in 6 out of 23 patients with one-variant in the PRKN gene (26.1%). In contrast, no additional variants were found in the 12 patients with a heterozygous variant in the PINK1 gene (0%) (Table 2). Notably, in the case of PRKN-31, long-read sequencing with 1 SV caller (Severus) detected a deletion in exon 3, a finding not reported by the other 2 SV callers, Sniffles and CuteSV. However, manual inspection using IGV revealed that this was a deletion spanning exons 3 and 4, underscoring the importance of manual curation (Fig S2).
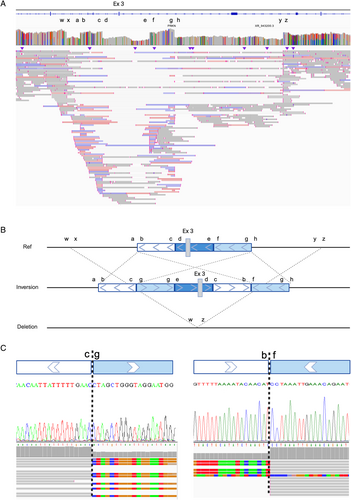
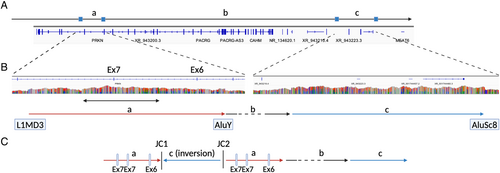
In the 6 “new” two-variant PRKN cases identified only by long-read sequencing, 1 case had inversion, 3 showed overlapping pathogenic PRKN SVs in each allele, and 2 cases carried a duplication (Table 3). Specifically, a complex inversion including exon 3 was identified from a patient (PRKN-10) previously recognized to have an exon 3 deletion via MLPA (Fig 2). All SV callers identified 2 overlapping inversions in the same region, including exon 3. Detailed analysis using the IGV by linking mapped reads revealed an inversion involving the region of exon 3, along with duplication of the flanking regions on both sides of exon 3 (Fig 2A,B). Additionally, we also confirmed the known exon 3 deletion in another allele by IGV. To confirm the inversion and deletion, we amplified the sequence of breakpoints and performed Sanger sequencing to ensure that the sequences surrounding the breakpoints were the same for long-read sequencing and Sanger sequencing (Fig 2C). This variant appeared to be an inversion of region including exon 3 accompanied by the duplications of flanking regions.
| Variants by MLPA and short-read seq | Variant one by long-read seq | Variant two by long-read seq | Notes | |
|---|---|---|---|---|
| PRKN-10 | PRKN exon 3 deletion | PRKN exon 3 deletion | PRKN exon 3 complex inversion | Complex structural variant |
| PRKN-11 | PRKN exon 2 deletion | PRKN exon 2 deletion and exon 3–4 duplication | PRKN exon 4 deletion | Deletion and duplication overlap |
| PRKN-21 | PRKN c.535-3A>G(T>C)(p.G179RfsX10) | PRKN c.535-3A>G(p.G179RfsX10) | PRKN exon 6 duplication | Duplication |
| PRKN-24 | PRKN exon 5–6 duplication | PRKN exon 6 duplication | PRKN exon 5–6 duplication | Duplication overlap |
| PRKN-31 | PRKN exon 2–3 deletion | PRKN exon 3 deletion | PRKN exon 2 deletion | Adjacent exon deletion on the different alleles |
| PRKN-34 | PRKN c.536delG_p.G179Vfs*9 | PRKN c.536delG(p.G179Vfs*9) | PRKN exon 6 duplication | Duplication |
- MLPA = multiplex ligation-dependent probe amplification; seq = sequencing.
Three subjects (PRKN-11, PRKN-24, and PRKN-31) presented with overlap of duplication and/or deletion in the same allelic exons, making it difficult to identify and judge by MLPA. PRKN-11, harboring exon 2 deletion identified by MLPA, was identified to have duplication of exon 3–4, exon 2 deletion, and exon 4 deletion (Fig S2). PEPPER-Margin-DeepVariant was not able to phase the variants around these exons, likely because of insufficient sequence length (N50). However, manual phasing determined that the duplication of exon 3–4 and exon 2 deletion were located on the same allele, whereas the exon 4 deletion was on another allele. PRKN-24 was identified to have exon 6 duplication and exon 5–6 duplication by long-read sequencing. In MLPA, because the 2 duplications overlapped, it is not possible to differentiate those two-variants (Fig S2). PRKN-31, who was identified to have exon 2–3 deletion by MLPA, appeared to have separate deletions of exon 2 and exon 3 deletion (Fig 3). The absence of a heterozygous variant between these deletions made phasing challenging, but the patient's phenotype suggests that the two-variants are unlikely to be on the same allele. Additionally, 2 cases (PRKN-21 and PRKN-34) carried duplications that were not detected by MLPA (Fig S2).
Alongside these duplications, PRKN-21 and PRKN-34 carried known pathogenic SNVs (c.535-3A>G(p.G179RfsX10) and c.536delG(p.G179Vfs*9)) that were identified by short-read targeted sequencing, which were also confirmed by long-read sequencing.
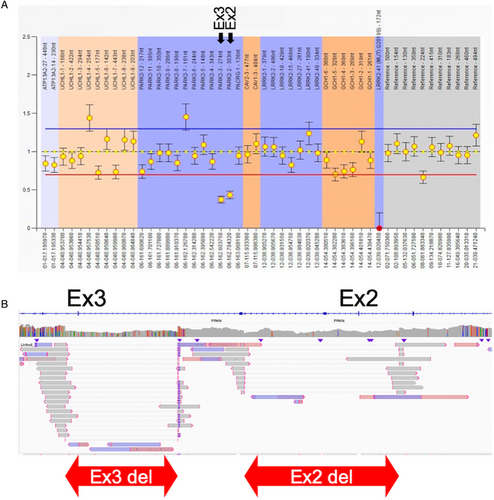
One case, PRKN-9, was characterized by a complex SV labeled as duplication-normal-inversion/duplication (DUP-NML-INV/DUP).31 DUP-NML-INV/DUP is a complex structural variant, which is caused by Alu-mediated rearrangements, strand dissociation followed by template switches during replication, which is reported in patients of Pelizaeus-Merzbacher disease (PLP1) and also in the CNV screening study of 17p13.3 but has not reported in PRKN locus before.32, 33 MLPA indicated that this patient carried an exon 7 duplication. However, long-read sequencing identified not only an exon 7 multiplication, but also an apparent increase of sequence reads overlapping with exon 6 and 7, suggesting the presence of an additional duplication (Fig 4). Further examination of split reads revealed a unique pattern. The reads mapped at 2 distinct breakpoints (Fig S3A–D) did not align with each other. Instead, they were found to align to a genomic region located 3 megabases (Mb) distant (Fig S3E–H). This pattern of alignment suggests the presence of a complex genomic variant, denoted as DUP-NML-INV/DUP, resulting in quadruplication of exon 4 and duplication of exon 3 (Fig 4). To validate this finding, we amplified the breakpoints junction (JC) 1 and JC 2 in Figure 4, which are unique to this individual. Through this, we confirmed that this set of breakpoints only exists in this individual, therefore, they are not present in control samples (Fig S4). All the variants identified by long-read sequencing in this study are summarized in Table S7.
Clinical Symptoms of Long-Read Diagnosed PRKN-PD
Clinical symptoms of the 6 “new” two-variants PRKN cases are summarized in Table S8. All patients except PRKN-25 presented typical presentations of PRKN-PD, showing AAO younger than 40 (AAO; 29.7 ± 14.84), normal heart-to-mediastinum (H/M) ratio in 123I-metaiodobenzylguanidine (MIBG) myocardial scintigraphy (75% [3/4]), less common autonomic symptoms (constipation 33% [2/6], urinary disturbance 0% [0/6], orthostatic hypotension 33% [2/6]), good response to levodopa (100% [6/6]), and less frequent olfactory dysfunction (0% [0/6]). PRKN-25, who harbored duplications of exon 5 and exon 5–6, had an AAO 49 and a family history of progressive supranuclear palsy (father). This patient had a decreased H/M ratio on MIBG myocardial scintigraphy, various autonomic symptoms, and levodopa equivalent dose of 1,300 mg 10 years after onset, which is atypical for PRKN-PD as it typically require low dose levodopa and does not show decreased H/M ratio.
We then compared the clinical features between PRKN two-variant carriers and PRKN one-variant carriers. Five features showed different trends between one- and two-variant carriers (AAO, disease duration, gait disturbance, dystonia showing response to levodopa, and dystonia at onset). For example, the age of onset was younger in two-variant carriers (one-variant vs two-variant; 38.0 ± 7.16 vs 28.6 ± 11.32, p value = 0.0064) (Tables S9 and S10, Fig S5).
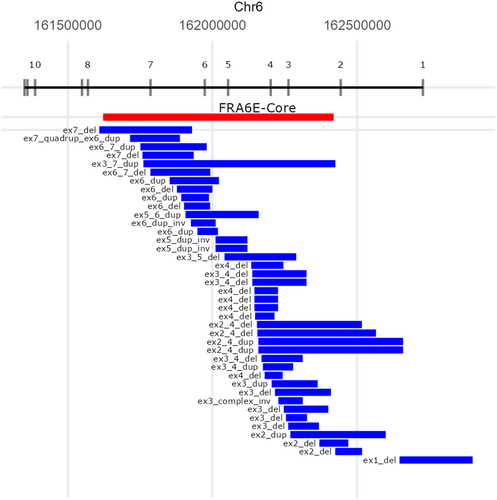
Breakpoints of Pathogenic SVs
All the breakpoints and the locations of pathogenic SVs of PRKN identified in this study are summarized in Figure 5 and Table S11. PRKN is located in one of the common fragile sites (CFS) in the genome, namely FRA6E, which makes the PRKN gene prone to have SVs. CFS are vulnerable to replication stress and often cause DNA breakage in this region, characterized by late replication, paucity of replication origins, and the ability to form DNA secondary structures.34 In addition, Figure 5 also presents the core of FRA6E, as defined by Denison et al.35 using BAC clones RPCI-1119H20 and RPCI-1179P19. Because the precise location of RPCI-1179P19 was unavailable, we used D6S1599 to represent the core visually.35 All identified pathogenic SVs, except for two-variants, had at least 1 breakpoint located in the core of FRA6E. (95%, 38/40) The 2 other variants were exon 1 deletion and exon 2 deletion of PRKN.
Discussion
In this study, we explored the performance of long-read sequencing for identifying SNV and complex SV in the PRKN/PINK1 genes. We included 12 known two-variants PRKN-PD as “positive controls” and all 24 variants were successfully identified. Next, we wanted to identify complex and previously undetected secondary variants in PRKN/PINK1 heterozygous carrier patients, potentially demonstrating that complex SVs and overlapping SVs of PRKN are likely missed by traditional sequencing methods and MLPA. In our cohort, we could identify a second variant in 26% (n = 6) of the PRKN heterozygous carriers and in 0% of the PINK1 heterozygous carriers. This study shows the utilization of long-read sequencing in the diagnosis of EOPD and long-read sequencing should be considered as a next step after short-read sequencing and MLPA for unresolved EOPD cases, approximately 5% to 10% of PD patients can be classified as monogenic PD, which means a single gene is mainly responsible for their disease development.2 In our study using short-read targeted resequencing for PD-related genes in the EOPD population, surprisingly, 60% of these patients remained undiagnosed. However, our research indicates that using long-read sequencing could be more effective. More than 20% of patients with a single variant in the PRKN gene were successfully diagnosed with this method in this study. Therefore, for those patients who remained undiagnosed after short-read sequencing, long-read sequencing might provide a diagnosis. Given that PRKN along with GBA1 and LRRK2, are targets for gene therapy in PD, applying long-read sequencing to the EOPD population may offer a broader range of candidates for the upcoming gene-therapy era.36 We anticipate that the utilization of long-read sequencing will become more widespread in the diagnosis of familial PD, particularly in unresolved high suspect monogenic/early onset cases.
One inversion was detected from 23 heterozygous PRKN carriers in the Japanese population. Considering the case of the massive inversion we recently reported, it is suggested that inversions in PRKN are not an extremely rare type of SV.17 We have also identified another complex SV, DUP-NML-INV/DUP, which included PRKN exons (Fig 4). For this variant, 2 of the junctions overlapped with an Alu transposable element in the reference genome, which is in line with the previous reports that described DUP-NML-INV/DUP was mediated by Alu-Alu rearrangements (Fig 3).33, 37, 38 This case underscores the complexity of SVs in PRKN.
These cases highlight the utility of long-read sequencing and we believe that long-read sequencing should be considered as a next step after short-read sequencing and MLPA for unresolved EOPD cases, especially with a heterozygous PRKN variant. Moreover, although it may not be frequent, we may need to consider that there should be EOPD cases with PRKN-PD phenotype harboring homozygous variants of complex SVs of PRKN when short read sequencing could identify any pathogenic variant as overlap of deletion and duplication in the same exact exon may be missed by MLPA.
In addition to this study, 3 cases of a pathogenic inversion involving PRKN have been reported.17, 39, 40 One case from the Yemenite-Jewish population describes EOPD patients from a consanguineous family with a homozygous 77kb inversion involving exon 5. Second, our case from Japan showed monozygotic twins with compound heterozygous PRKN variants of exon 3 deletion and 7Mb inversion including exon 1 to 11. The last case was from Poland, describing inversion including exon 2 to exon 5, which was a part of duplication. Given the observation of PRKN inversion across various populations, including Jewish, European, and Asian, it is reasonable to infer that this genetic variation can be identified in a wide range of ethnic backgrounds. We may need to sequence a larger number of samples from diverse populations to know the frequency of inversions of PRKN.
Long-read sequencing also helped identify the variants of PRKN when the SVs of each allele overlap or when the SVs are contiguous. Three of 6 long-read diagnosed PRKN-PD cases harbored overlapping or contiguous SVs. We have previously reported that overlapping of a deletion and duplication in the same allelic exon could be normal in qPCR and differentiated them using parental DNA.41 It is natural to consider that overlap of deletion and duplication can be missed by MLPA. In this study, we also identified overlap of deletion and duplication missed by MLPA (PRKN-11). Parental DNA was needed to phase the variants in conventional methods, however, using long-read sequencing, it is able to differentiate and phase the overlapped variants only by proband's DNA. Long-read sequencing was also valuable in distinguishing between 2 deletions in consecutive exons that appeared as a single deletion encompassing 2 exons in MLPA (PRKN-32). These findings underlined the utility of long-read sequencing in accurately diagnosing PRKN-PD when SVs are overlapped or continuous.
When we compared the clinical phenotype of PRKN two-variants group (n = 18) and PRKN single variant group (n = 18), 5 features had a significant correlation with the number of the PRKN variants (Table S10, Fig S5). AAO was younger in the two-variants group, suggesting that true PRKN-PD patients are likely to have younger AAO. Moreover, disease duration was shorter in the one-variant group. It may cause inaccurate diagnosis of PD in those patients.
In this study, we could not find any SVs in PINK1. Reported SVs of PINK1 include deletion of single exon, multiple exons, or whole gene in multiple populations and exonic duplications.16 We hypothesized that there may be complex SVs as a hidden variant, but none was identified. Complex SVs were not identified because they were truly absent or because of the small number of samples (n = 12). We plan to perform long-read sequencing on larger samples to further elucidate the presence of complex SVs in PINK1.
Approximately three-quarters of PRKN one-variant carriers and all PINK1 one-variant carriers remained genetically undiagnosed after long-read sequencing. Several possibilities could explain this situation, including deep intronic variants, repeat expansions, accumulation of somatic variants in PRKN or PINK1, or that neither PRKN nor PINK1 is the actual cause of disease. To confirm these possibilities, additional experiments are required, such as RNA sequencing of deep mitochondrial sequencing from heterozygous PRKN/PINK1 carriers after long-read sequencing. We plan to expand our research to identify other hidden factors in heterozygous PRKN/PINK1 variant carriers that contribute to the development of PD.
A key question that can be addressed from the findings of this study is: when do we need to consider long-read sequencing for EOPD? An important consideration is that MLPA is generally more affordable compared to long-read sequencing, which allows MLPA to expand the study population.42 It is reasonable, therefore, to perform MLPA to screen for SVs first.42 In our previous study, we combined MLPA with targeted resequencing/Sanger sequencing to analyze EOPD patients (n = 918) with an AAO younger than 50 years in a Japanese cohort. We identified that 6.4% of the patients harbored two-variants in the PRKN gene, whereas 3.9% presented with a single variant.11 A study from the United Kingdom reported 2.3% of two-variant carriers of PRKN and 3.8% of single variant carriers from EOPD with AAO younger than 50 using direct sequencing and MLPA.43 In addition, a recent paper showed PRKN-PD is more common (18 per 100,000 individuals) than it has been thought (35,000–70,000 worldwide), which suggests the number of PD patients with PRKN variant should be larger.44, 45 Therefore, it is assumed that there is a certain number of EOPD patients with heterozygous PRKN variants after checking pathogenic SNVs and CNVs by conventional methods, which is considered to be a good application for long-read sequencing, as we did in this study.
To date, long-read sequencing is not generally available in clinical testing mainly because of its cost. Adaptive sampling in Oxford nanopore may help to decrease the cost because it enables sequencing 4 to 5 DNA samples per 1 flow cell for a selected region of interest. We expect that the cost of sequencing will decrease and that we and others can apply this method to larger collections of EOPD cases and families.
Several studies have demonstrated the utility of long-read sequencing in PD research. The GBA1 gene, a well-known risk factor for PD, presents a particular challenge for variant calling because of the presence of its pseudogene, GBAP1, which shares 96% sequence similarity with GBA1, along with the gene's complex recombination events. Two studies have highlighted the efficacy of long-read sequencing in addressing this challenge.46, 47 Additionally, Tseng et al48 used PacBio long-read sequencing to characterize novel transcripts in the SNCA gene region. Our research also underscores the potential of long-read sequencing in resolving complex genomic regions associated with PD, and we anticipate further studies will continue to advance understanding in these challenging areas.
Our study may influence the understanding of the significance of PRKN heterozygous variants in the onset of PD. The role of PRKN heterozygous variants in PD remains controversial.8-11, 13-15 However, previous studies investigating the effect of PRKN heterozygous variants did not use long-read sequencing for variant screening. Given that SVs can be missed by MLPA and short-read targeted sequencing, it is plausible that the observed association of PRKN heterozygous variants with PD may be driven by undetected SVs.
To our knowledge, this is the first large-scale study to apply long-read sequencing to PRKN SVs to describe the breakpoints of pathogenic SVs more accurately. Notably, almost all PRKN SVs we identified in this study were located in the central core of FRA6E (Fig 5).35 When we compared the location of SVs identified in this study to the SVs recurrently observed in the study from Mitsui et al,49 4 SVs were common. These 4 SVs were found from Japanese or Asian populations in their study, but not from European populations. These facts support the necessity of long-read sequencing for the identification of complex SVs, because it seems to be difficult to identify a region in which complex SVs frequently occur and screen them using cheaper techniques like Sanger sequencing. Moreover, as we confirmed that pathogenic SVs of PRKN were concentrated in the FRA6E core. We speculate that looking closer to the SVs in common fragile sites may lead us to identify more disease or phenotype-related SVs in neurodegenerative disease, especially in familial cases.
This study has some limitations. First, although the current sample size for PRKN-PD is relatively large, it remains limited for accurately determining the true frequency of complex structural variants, such as inversions. More long-read sequencing data, including large numbers of controls, is needed to know the frequency of inversions across populations. Second, we were not able to confirm the changes in RNA transcripts in samples with complex SVs in PRKN. Attempts using reverse transcription-PCR and RNA sequencing from mRNA extracted from peripheral blood were unsuccessful because of the low expression of PRKN, and unfortunately, no other patient material was available. Third, we only had access to samples from Japanese ancestry. A more diverse population is needed to know the true significance of complex SVs in EOPD. We are now in the process of applying long-read sequencing to different ancestral populations.
In summary, this study was the first study to use long-read sequencing on a large group of EOPD patients to identify hidden and complex SVs. This study demonstrated the complexity of SVs in the PRKN gene, which is even more complicated than previously thought. Additionally, the study highlighted the effectiveness of long-read sequencing in researching the genetics of EOPD. It is expected that the application of long-read sequencing will increase, leading to more accurate and faster diagnoses, which is important for PRKN-PD given the potential need for genetic counseling, different progression versus idiopathic PD, and eligibility for clinical trials.
Acknowledgments
We thank the Biowulf team, as this study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH) (http://hpc.nih.gov). This work was in part supported by the Intractable Disease Research Center of Juntendo University Graduate School of Medicine. This research was funded in part by the Intramural Research Program of the NIH, National Institute on Aging, NIH (grant numbers: 1ZIAAG000542-01 and 1ZIAAG000538-04), the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers: 24K02372 and 23K06958, M.F.; 22K07542, H.Y.; 21K07283,Y.L.; 20K07893, K.N.; 21H04820 and 24H00068, N.H.), the Japan Science and Technology Agency Moonshot R&D Program (grant number: JPMJMS2024-5, N.H.), AMED (grant number: 23bm1423015h0001, M.F. and N.H. and 24ek0109677h0002, N.H.), Subsidies for Current Expenditures to Private Institutions of Higher Education from the Promotion and Mutual Aid Corporation for Private Schools of Japan, through a subaward from Juntendo University (for M.F. and N.H.), and the Research Institute for Diseases of Old Age, Juntendo University Graduate School of Medicine (for M.F., Y.H., and N.H.). K.D. was supported by JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH. We thank all the participants who contributed to this study. Figures 1, 2, and 4 were generated on www.biorender.com.
Author Contributions
K.D., M.F., C.B. and N.H. contributed to the conception and design of the study; K.D., H.Y., L.M., B.B., R.G., K.P., M.I., M.F., Y.I., K.N., S.M., M.H., K.T., K.J.B., M.F., and C.B. contributed to the acquisition and analysis of data; K.D., K.J.B., M.F., C.B., and N.H. contributed to drafting of the manuscript and figures.
Potential Conflicts of Interest
Nothing to report.
Open Research
Data Availability
The raw data supporting the findings of this study unfortunately cannot be made publicly available because of local ethical regulations. The data are available from the corresponding author on reasonable request and by implementing a material transfer agreement.