Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
Issue Information
- First Published: 23 February 2023

Metabarcoding Silk Road. The cover reveals the combination of environmental DNA (eDNA) and metabarcoding, a new approach for large scale plant diversity assessment and conservation developed by Liu et al, describing the plant diversity in the east of Modern Silk Road. The warm color represents the extremely dry climate of Modern Silk Road, the double helix structures and plants on the ground represent the eDNA in soil and the plant species diversity, respectively. The Silk Road indicates a long way for metabarcoding in the field, which requires counterparts from various countries to carry out more diverse and extensive metabarcoding researches. Designed by Yanlei Liu, & Paper Artist.
EDITORIAL
iMeta progress and acknowledgment of reviewers in 2022
- First Published: 13 February 2023
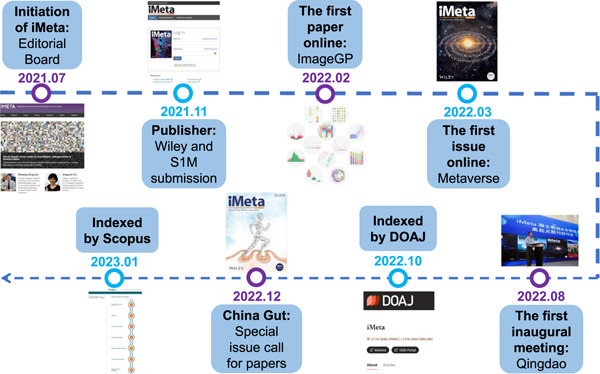
Milestones of the first year of iMeta. iMeta is an open-access Wiley partner journal launched by iMeta Science Society consisting of worldwide scientists in bioinformatics and metagenomics. In 2022, iMeta released four issues, including 60 publications with a total of 340 citations. iMeta has been indexed in several databases, including Google Scholar, Crossref, CNKI, Dimensions, PubMed (partial), DOAJ, and Scopus. Thanks to the editorial board members and reviewers for their contributions to the iMeta in 2022.
SHORT COMMUNICATION
Warmer and drier ecosystems select for smaller bacterial genomes in global soils
- First Published: 03 January 2023
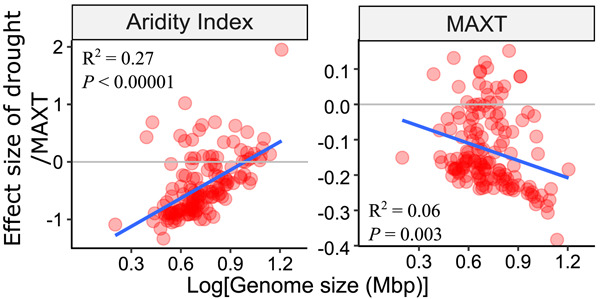
We used surface soils collected from 237 sites across the globe and analyzed how environmental conditions affect soil bacterial occurrences and genome size. We used a joint species distribution model to quantify the effects of environments on species occurrences and found that aridity was a major regulator of genome size with warmer and drier environments selecting bacteria with smaller genomes.
RESEARCH ARTICLES
Deciphering factors driving soil microbial life-history strategies in restored grasslands
- First Published: 04 December 2022
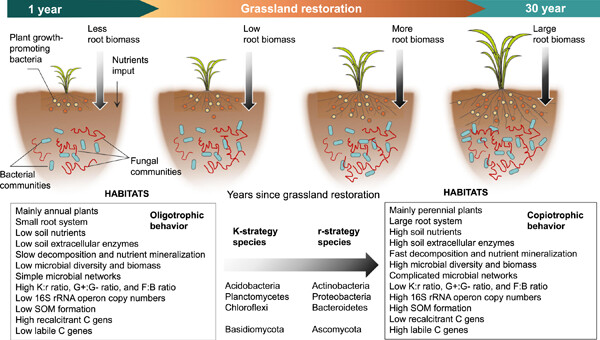
We concluded that the macroecological theory could be applied to soil microbial life-history traits and also highlight the key effects of plant and soil properties on the changes in microbial strategies. These findings were suitable for this Journal (significantly advance the field of Microbial Ecology) and first proved that macroecological theory could be applied to soil microbial community, which was benefit to the development of microbial ecology and can be regarded as the evidence for most of researches.
Pan-cancer analyses reveal molecular and clinical characteristics of cuproptosis regulators
- First Published: 05 December 2022
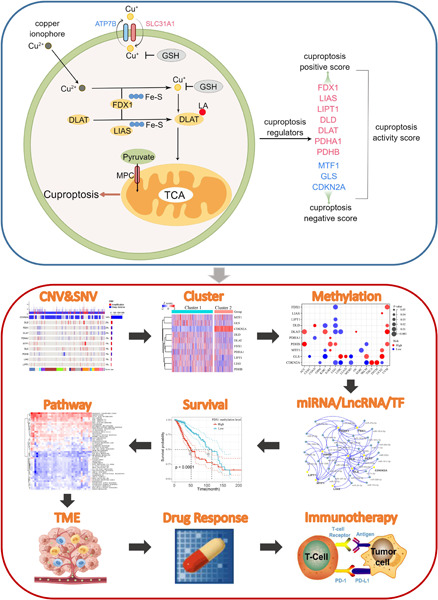
We developed a cuproptosis activity score to reflect the overall cuproptosis level, which could provide a methodological reference for future cuproptosis studies. Cuproptosis is associated with a variety of tumor-related pathways, and in particular, tumors with high fatty acid metabolism may be more sensitive to cuproptosis. Cuproptosis is associated with the regulation of the tumor microenvironment and can predict the outcome of immunotherapy patients based on cuproptosis activity. This study identified cuproptosis-related microRNAs, long noncoding RNAs, and transcription factors in each tumor type, which can provide a resource and reference for future cuproptosis-related research.
What determines plant species diversity along the Modern Silk Road in the east?
- First Published: 09 January 2023

Environmental DNA (eDNA) is a quick and accurate method for biodiversity assessment. This study first used eDNA-assessed regional plant species diversity. Temperature instead of precipitation is the main factors affecting plant diversity. Human activities were proven to affect plant species diversity in desert areas. More instant plant diversity results need to be done for desertification control.
Ecological dynamics imposes fundamental challenges in community-based microbial source tracking
- First Published: 05 January 2023
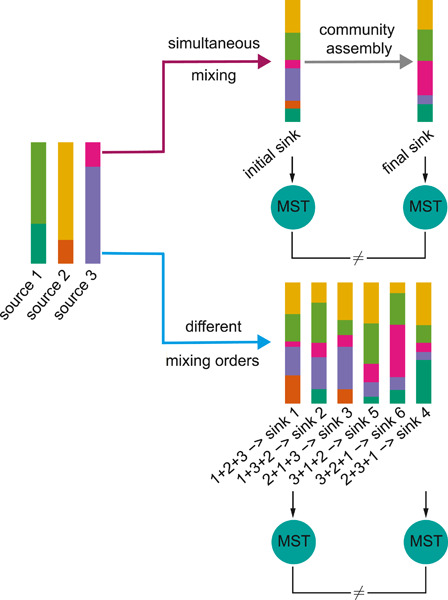
Quantifying the contributions of possible environmental sources (“sources”) to a specific microbial community (“sink”) is a classical problem in microbiology known as microbial source tracking (MST). We show compelling evidence that solving the MST problem using existing MST solvers is impractical when ecological dynamics plays a role in community assembly. In particular, we clearly demonstrate that the presence of either microbial interactions or priority effects will render the MST problem mathematically unsolvable for MST solvers.
Destabilized microbial networks with distinct performances of abundant and rare biospheres in maintaining networks under increasing salinity stress
- First Published: 09 January 2023
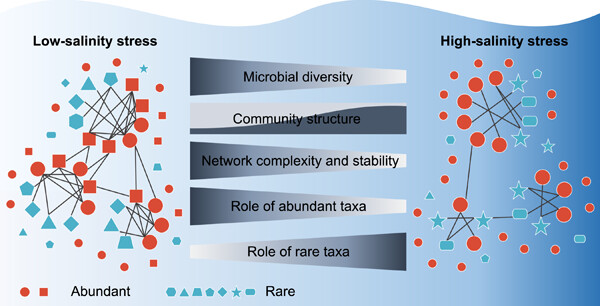
This study revealed that, with increasing salinity, the microbial diversity declined, the taxonomic structure underwent marked turnover, and the complexity and stability of ecological networks diminished. In low-stress conditions, the abundant biosphere played a more important role than the rare biosphere in stabilizing ecological networks, but the difference between their relative importance narrowed significantly with the increasing stress.
Synergistic effects of diazotrophs and arbuscular mycorrhizal fungi on soil biological nitrogen fixation after three decades of fertilization
- First Published: 27 January 2023
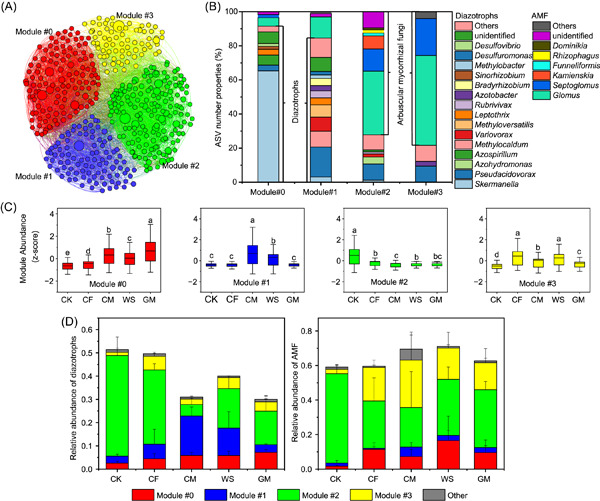
In this study, a 33-year fertilization experiment was conducted to investigate the changes in soil BNF rates, diazotrophic and AMF communities, and their interactions after various long-term fertilizations. We found a remarkable increase in soil BNF rates after more than three decades of fertilization compared with nonfertilized soil, and the green manure treatment rendered the highest enhancement.
MBPD: A multiple bacterial pathogen detection pipeline for One Health practices
- First Published: 31 January 2023
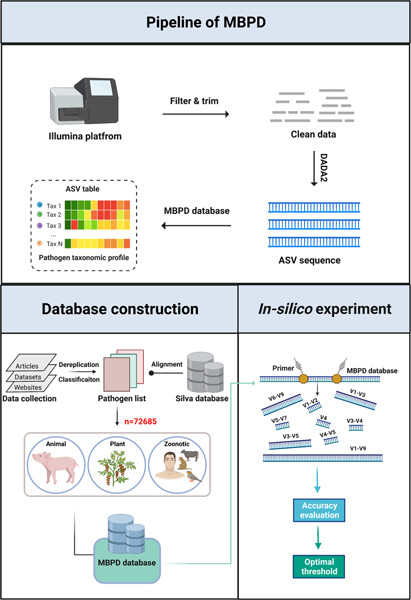
Multiple bacterial pathogen detection (MBPD) provides the accurate and comprehensive detection of bacterial pathogens from biological and environmental samples based on 16S rRNA gene sequencing. By constructing a relatively complete database of 72,685 sequences for wide identification across a broad range of bacterial pathogens causing animal, plant, and zoonotic diseases, MBPD can provide the appropriate threshold for common variable regions of 16S rRNA gene in taxonomy alignment based on the recommendation of in silico experiment. MBPD is freely available at https://github.com/LorMeBioAI/MBPDLorMeBioAI/MBPD.
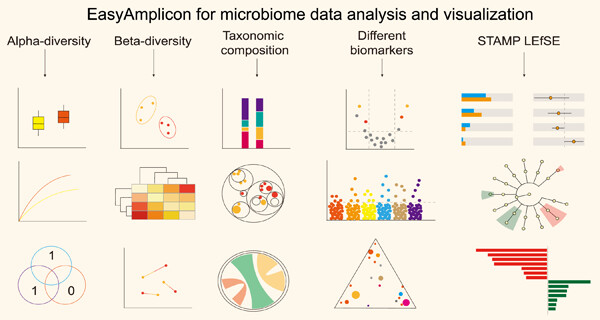
EasyAmplicon: An easy-to-use, open-source, reproducible, and community-based pipeline for amplicon data analysis in microbiome research
- First Published: 27 January 2023
REVIEW ARTICLES
New insights into the mechanisms of high-fat diet mediated gut microbiota in chronic diseases
- First Published: 05 January 2023
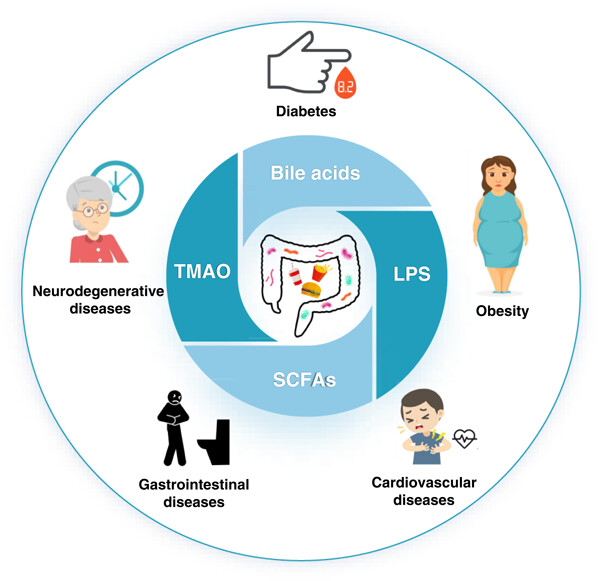
Gut microbiota dysbiosis increases the host's susceptibility to diseases. Identified characteristic microbes in high-fat diet (HFD) induced obesity, diabetes, gastrointestinal diseases, neurodegeneration, and cardiovascular diseases. Bile acids, lipopolysaccharide, short-chain fatty acids, and trimethylamine N-oxide are the commonalities in HFD-induced chronic diseases. FXR, TGR5, NF-κB, PPAR-γ, and PERK signaling were critical for the interacted mechanisms toward microbiota modulation. Provided new insights into mechanisms among microbiota, metabolites, and immune responses in chronic diseases.
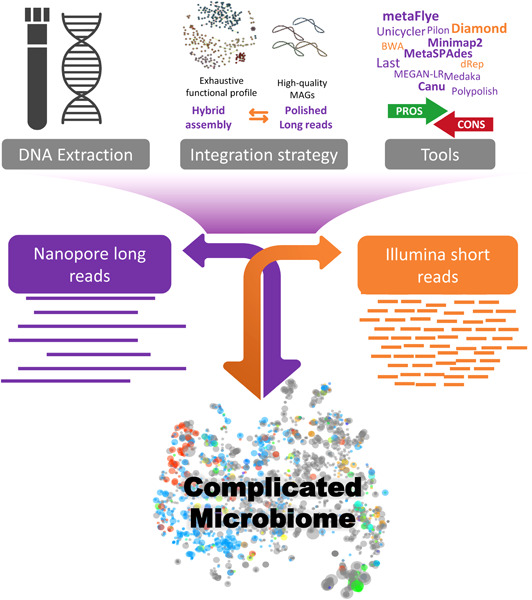
Strategies and tools in illumina and nanopore-integrated metagenomic analysis of microbiome data
- First Published: 09 January 2023
Applying multi-omics toward tumor microbiome research
- First Published: 09 January 2023
Decoding the role of immune T cells: A new territory for improvement of metabolic-associated fatty liver disease
- First Published: 18 January 2023
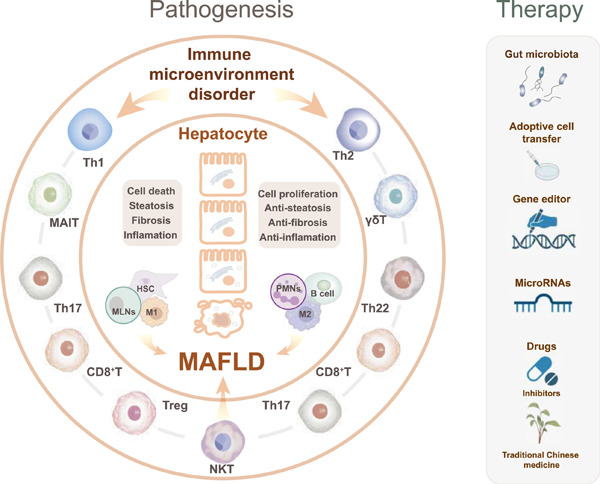
Immune T cells are involved in metabolic-associated fatty liver disease (MAFLD) progression either in a positive or negative manner and the therapeutic methods of MAFLD caused by disordered T cells include gut microbiota regulation, adoptive cell transfer, gene editor microRNAs, drugs (inhibitors/traditional Chinese medicine) therapy.
PROTOCOL
Using PhyloSuite for molecular phylogeny and tree-based analyses
- First Published: 16 February 2023
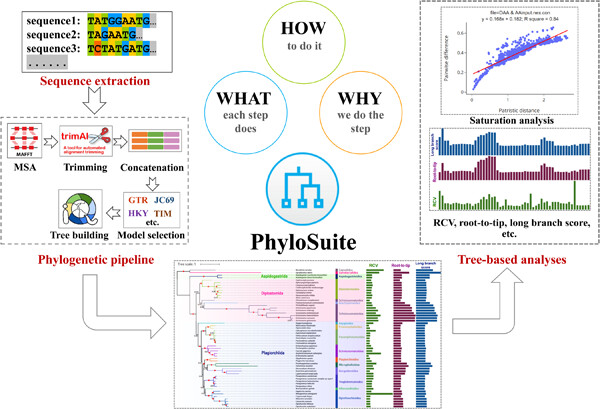
A new release of PhyloSuite, capable of conducting tree-based analyses. Detailed guidelines for each step of phylogenetic and tree-based analyses, following the “What? Why? and How?” structure. This protocol will help beginners learn how to conduct multilocus phylogenetic analyses and help experienced scientists improve their efficiency.
COMMENTARY
A guide for comparing microbial co-occurrence networks
- First Published: 10 January 2023
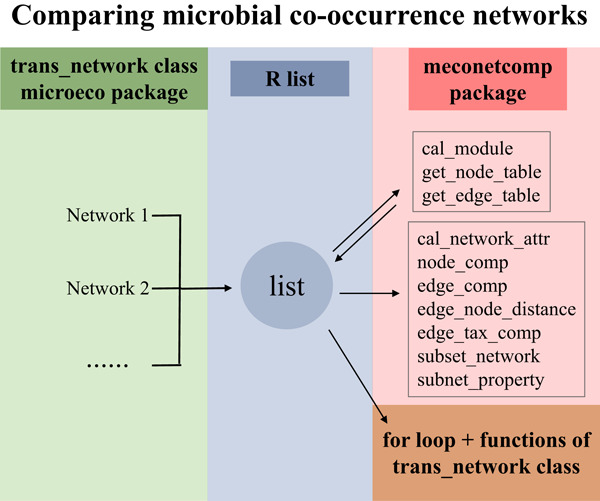
The article provides a pipeline for comparing microbial co-occurrence networks based on the R microeco package and meconetcomp package. It has high flexibility and expansibility and can help users efficiently compare networks built from different groups of samples or different construction approaches.
Metagenomic sequencing combined with flow cytometry facilitated a novel microbial risk assessment framework for bacterial pathogens in municipal wastewater without cultivation
- First Published: 05 January 2023
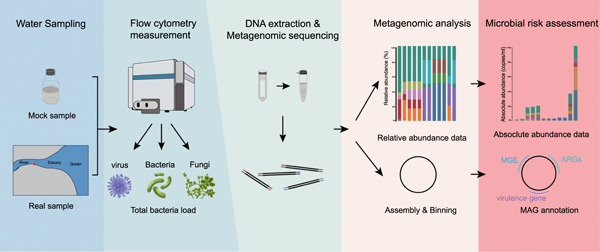
A workflow that combined metagenomic sequencing with flow cytometry was developed. The absolute abundance of pathogens was accurately estimated in mock communities and real samples. Metagenome-assembled genomes binned from metagenomic data set is robust in phylogenetic analysis and virulence profiling.
QMD: A new method to quantify microbial absolute abundance differences between groups
- First Published: 05 January 2023
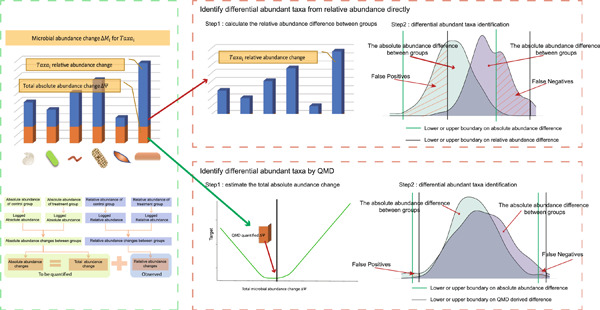
A new method, quantification of microbial absolute abundance differences (QMD), was proposed to estimate the microbial absolute abundance changes of each taxon under different conditions based on the microbial relative abundance. Compared with other methods, QMD displayed greater confidence in understanding microbiome dynamics between groups. We also provide QMD software to investigate common deviations and achieve a better understanding of microbiota changes under different conditions.
OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication
- First Published: 06 February 2023

OmicStudio focuses on speed, quality together with flexibility. Generally, OmicStudio can not only meet the users' demand of ordinary bioinformatics data analysis, statistics, and visualization, but also provides them freedom of data mining beyond developer's framework. Additionally, unlimited to developer's aesthetics, users can get more elegant graphs through customizing. Available online https://www.omicstudio.cn.
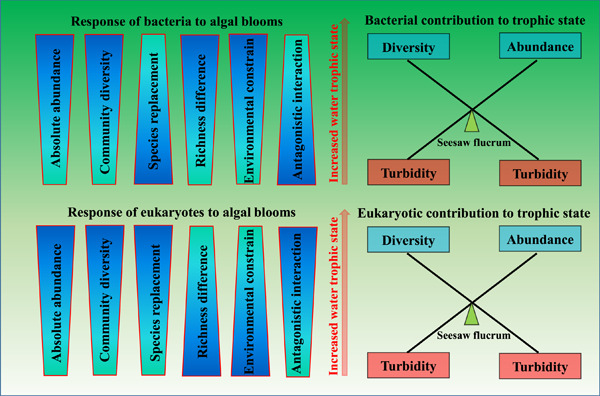
Differentiation strategies for planktonic bacteria and eukaryotes in response to aggravated algal blooms in urban lakes
- First Published: 31 January 2023
Plant colonization mediates the microbial community dynamics in glacier forelands of the Tibetan Plateau
- First Published: 14 February 2023
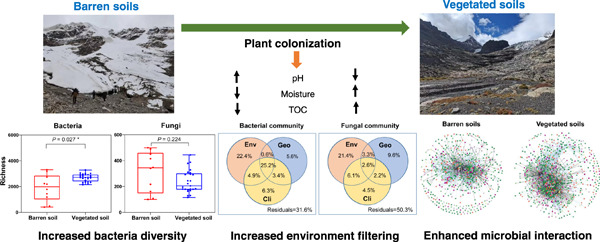
It has long been recognized that pH mediates community structure changes in glacier foreland soils. Here, we showed that pH changes resulted from plant colonization. Plant colonization reduced pH and increased soil organic carbon, which increased bacterial diversity, changed the community structure of both bacteria and fungi, enhanced environmental filtering, and improved microbial network disturbance resistance.








3066-988X.cover.jpg)