Journal list menu
Export Citations
Download PDFs
Front Cover
Acinetobacter baumannii has a novel β-ketoacyl-acyl carrier protein synthase III (KAS III), which can feed long, host-derived fatty acyl-CoA molecules into de novo fatty acid synthesis. The crystal structure of KAS III reveals structural changes in the active site that allow accommodation of octanoyl-CoA. For details, see the article by Lee et al. on pp. 567–577 of this issue.
- Page: i
- First Published: 28 May 2018
Issue Information
MicroCommentary
For Cryptococcus neoformans, responding to the copper status in a colonization niche is not just about copper
- Pages: 463-466
- First Published: 06 April 2018
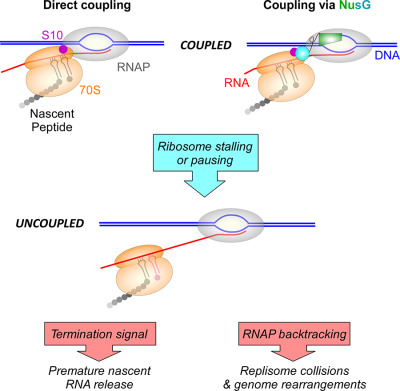
Rebuilding the bridge between transcription and translation
- Pages: 467-472
- First Published: 02 April 2018
Research Articles
Genome-wide analysis of the regulation of Cu metabolism in Cryptococcus neoformans
- Pages: 473-494
- First Published: 02 April 2018
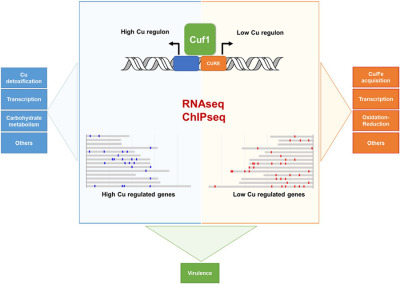
The essential yet toxic nature of Cu ions in living cells requires exquisite control of Cu homeostasis. The fungal pathogen C. neoformans regulates Cu homeostasis for survival during its complex host colonization process. During pulmonary infection host innate immune cells use Cu to kill C. neoformans, which responds by activating expression of Cu detoxifying proteins. However, during brain colonization, expression of the fungal Cu import machinery is activated and required for virulence. To achieve the genetic plasticity required for adaptation to a continuum of distinct Cu environments within the host, C. neoformans utilizes the Cu-responsive transcription factor, Cuf1. Cuf1 is unique as it senses and responds to both high and low Cu environments, activating different sets of genes dependent on environmental Cu status. Cells lacking Cuf1 are compromised for colonization of the lungs and brain, highlighting Cuf1 as an important virulence factor. A genome-wide assessment of Cuf1 binding sites and Cuf1-dependent transcription changes driven by Cu status identified novel genes required for adaptation to differential Cu environments. These genes and their regulation provide new insights into the adaptive responses to changes in host Cu availability and may reveal new targets for therapeutic intervention in cryptococcosis.
RNA-Seq and ChIP-Seq experiments identified the Cuf1-dependent Cryptococcus neoformans Cu regulon. Cuf1 regulation is intrinsically complex, Cuf1 functions as an activator, a repressor and potentially as a modulator of transcription in response to Cu availability. Cu- and Cuf1-dependent regulon in C. neoformans reveals new adaptive mechanisms for Cu homeostasis, and identifies potential pathogen-specific targets for therapeutic intervention.
Escherichia coli transcription factor NusG binds to 70S ribosomes
- Pages: 495-504
- First Published: 25 March 2018
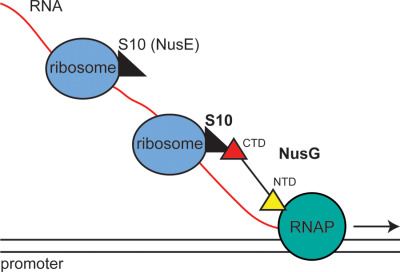
NusG binds S10 (NusE) in vitro and in vivo. NusG thus links the lead ribosome to elongating RNA polymerase coupling transcription with translation. Mutations in the S10 (NusE) interface with NusG allow RNAP to continue to transcribe when translation elongation is arrested, leading to RNAP - replisome clashes.
Insights into transcriptional silencing and anti-silencing in Shigella flexneri: a detailed molecular analysis of the icsP virulence locus
- Pages: 505-518
- First Published: 17 February 2018
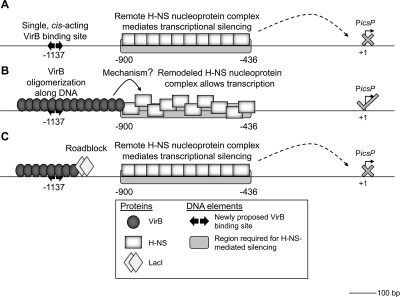
Remote regulatory elements are required for H-NS-mediated transcriptional silencing and VirB-dependent anti-silencing of icsP. A: Region required for transcriptional silencing displays high affinity for H-NS. H-NS bridging between this site and promoter regions is not supported. B: A single, cis–acting, VirB binding site is required for transcriptional anti-silencing and allows bi-directional oligomerization of VirB from this site in vitro. C: LacI protein blocks anti-silencing by VirB when positioned downstream of the VirB binding site.
A Toxoplasma gondii locus required for the direct manipulation of host mitochondria has maintained multiple ancestral functions
- Pages: 519-535
- First Published: 05 March 2018
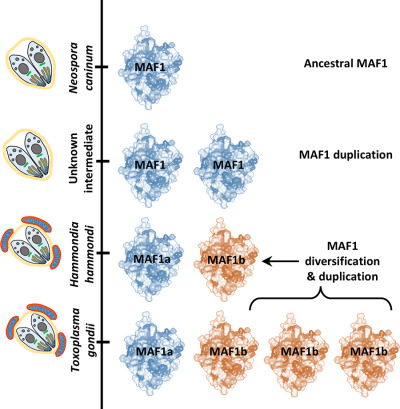
Only some mitochondrial association factor 1 (MAF1) paralogs in Toxoplasma gondii mediate association between the vacuole and host mitochondria. Using comparative structural analyses we find that functional differences are determined by only a few residues in the C-terminus. We also show that a conserved vestigial ADPr binding cleft may represent an ancestral function that was likely present in an ancestral MAF1 ortholog that had yet to acquire the ability to manipulate host mitochondria during infection.
Tandem tyrosine phosphosites in the Enteropathogenic Escherichia coli chaperone CesT are required for differential type III effector translocation and virulence
- Pages: 536-550
- First Published: 06 March 2018
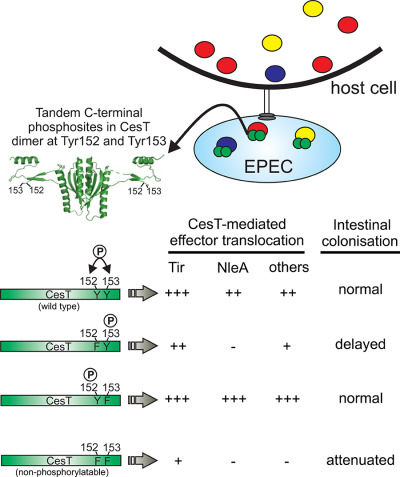
Pathogens with type III secretion systems (T3SS) absolutely require dedicated chaperone proteins to mediate efficient effector protein injection into host cells during infection. In this study, we implicate tandem tyrosine phosphosites of CesT in differentially regulating effector secretion and contributing to intestinal disease. These findings expand the functional importance of CesT in context of bacterial pathogenesis, revealing an unexpected phosphosite specificity for a T3SS chaperone.
SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron
- Pages: 551-566
- First Published: 12 March 2018
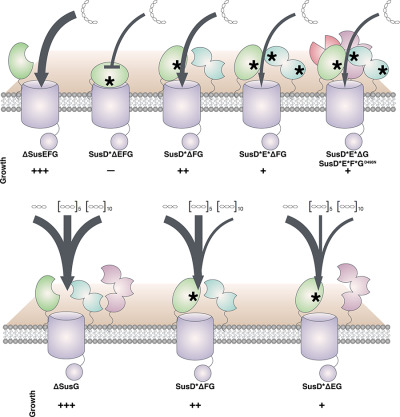
Human gut Bacteroidetes use multi-protein cell surface complexes to catabolize numerous dietary polysaccharides that the host cannot digest. Here, we reveal that starch-binding proteins of the starch utilization system in Bacteroides thetaiotaomicron facilitate glycan import independent of their binding sites and influence the length of glycan preferentially imported by the bacterium.
Structure and substrate specificity of β-ketoacyl-acyl carrier protein synthase III from Acinetobacter baumannii
- Pages: 567-577
- First Published: 12 March 2018
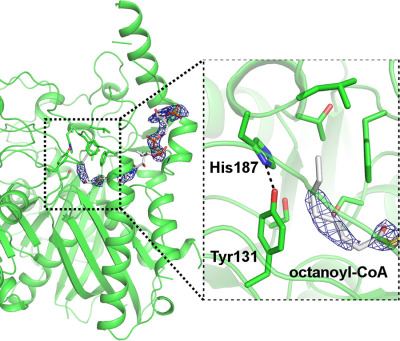
Acinetobacter baumannii has a novel β-ketoacyl-acyl carrier protein synthase III (KAS III), which can accept longer fatty acyl-CoAs and can feed into de novo fatty acid synthesis. This feature is clinically important as hosts' free fatty acids can be acquired and utilized by pathogens. The crystal structure of A. baumannii KAS III reveals a network of hydrogen bonds (His291-Asn314) and substitution of residues in the active site cavity to accommodate octanoyl-CoA.
Glutamine-rich toxic proteins GrtA, GrtB and GrtC together with the antisense RNA AsgR constitute a toxin–antitoxin-like system in Corynebacterium glutamicum
- Pages: 578-594
- First Published: 14 March 2018
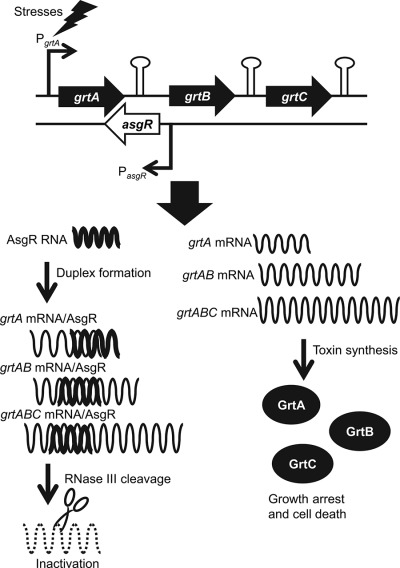
We report a new type I toxin-antitoxin system in Corynebacterium glutamicum which is a workhorse of amino acid production. The TA system is composed of three repeated glutamine-rich proteins encoded by grtABC genes and an antisense small RNA AsgR. The expression of grtABC is induced by several stressors while the repression of grtABC is achieved by the grtA-AsgR duplex formation at the 3′ untranslated region (3′-UTR) and subsequent degradation by RNase III.