Escherichia coli transcription factor NusG binds to 70S ribosomes
Summary
Transcription and translation are coupled processes in bacteria. A role of transcription elongation cofactor NusG in coupling has been suggested by in vitro structural studies. NMR revealed association of the NusG carboxy-terminal domain with S10 (NusE), implying a direct role for NusG as a bridge linking RNAP and the lead ribosome. Here we present the first in vitro and in vivo evidence of full-length NusG association with mature 70S ribosomes. Binding did not require accessory factors in vitro. Mutating the NusG:S10 binding interface at NusG F165 or NusE M88 and D97 residues weakened NusG:S10 association in vivo and completely abolished it in vitro, supporting the specificity of this interaction. Mutations in the binding interface increased sensitivity to chloramphenicol. This phenotype was suppressed by rpoB*35, an RNAP mutation that reduces replisome-RNAP clashes. We propose that weakened NusG:S10 interaction leads to uncoupling when translation is inhibited, with resulting RNAP backtracking, replication blocks and formation of lethal DNA double-strand breaks.
Graphical Abstract
NusG binds S10 (NusE) in vitro and in vivo. NusG thus links the lead ribosome to elongating RNA polymerase coupling transcription with translation. Mutations in the S10 (NusE) interface with NusG allow RNAP to continue to transcribe when translation elongation is arrested, leading to RNAP - replisome clashes.
Introduction
Coordinating the processes of transcription and translation is critical for cellular survival. In bacteria, newly transcribed mRNA is immediately translated by ribosomes trailing after RNAP. Uncoupling of transcription from translation induces attenuation and/or Rho-dependent polarity beyond nonsense codons within certain genes and operons (Adhya and Gottesman, 1978; Yanofsky, 1981; Richardson, 1991). Proshkin et al. (2010) demonstrated this coupling by quantifying the kinetic correlation between transcription and translation rates in vivo. Interaction between elongating RNAP and translating ribosomes was shown later in an in vitro system developed from purified components (Castro-Roa and Zenkin, 2012).
Escherichia coli transcription factor NusG (EcNusG) plays paradoxical roles in transcription regulation. It enhances RNAP processivity by suppressing pausing and preventing backtracking (Herbert et al., 2010). Although it accelerates transcription elongation, E. coli NusG stimulates Rho-dependent transcription termination both in vivo and in vitro (Sullivan and Gottesman, 1992; Li et al., 1993; Nehrke and Platt, 1994). In conjunction with NusB, NusE (S10) and NusA, NusG promotes antitermination in rrn operons (Li et al., 1992; Zellars and Squires, 1999; Torres et al., 2004). With λ N and the other Nus factors, NusG supports transcription antitermination on the phage λ chromosome. In B. subtilis however, NusG increases rather than reduces pausing (Yakhnin et al., 2008). Note that unlike EcNusG, BsNusG does not enhance Rho-dependent termination and is not essential for viability (Ingham et al., 1999).
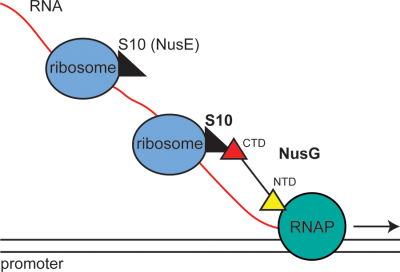
NMR studies revealed a complex between the NusG carboxy-terminal domain (NusG-CTD) and NusB-S10 (Burmann et al., 2010). Since the NusG N-terminal domain (NusG-NTD) interacts with RNAP (Mooney et al., 2009b), it was apparent that NusG might act as a molecular bridge coupling RNAP with the lead ribosome, i.e., coupling transcription with translation. Furthermore, the NusG-CTD interface that interacts with Rho protein to stimulate termination (Chalissery et al., 2011) is also bound by ribosomal protein S10. Therefore, NusG is not expected to enhance Rho-dependent transcription termination within translated genes.
Extrapolating from the studies of Burmann et al. (2010), we attempted to demonstrate a physical association between full-length NusG and mature 70S ribosomes both in vivo and in vitro, hence clarifying the role of NusG in coupling transcription with translation in bacteria. Our results show specific in vivo and in vitro association between NusG and S10 protein within the mature 70S ribosome. We found that the interaction interface of NusG and the ribosome complex is the same as that between NusG-CTD and S10 in the antitermination complex reported by Burmann et al. (2010). We also show that mutations in the NusG and S10 interface predicted to uncouple transcription from translation induce sensitivity to chloramphenicol. This sensitivity is suppressed by rpoB*35, an RNAP mutation that reduces replisome-transcription clashes (Trautinger et al., 2005).
Results
NusG and 70S ribosomes associate in vitro without accessory factors
Burmann et al. (2010) showed that a NusB:S10 complex associated with NusG-CTD in vitro. S10 also forms an integral part of the 30S ribosomal subunit. In their study Burmann et al. (2010) proposed a model for coupled transcription and translation in bacteria whereby NusG, which also binds RNAP, forms a bridge between the lead ribosome and RNAP. This stabilizes a coupled state and, additionally, blocks Rho-dependent termination of the elongating transcription complex. In accordance with this hypothesis, NusG protein should associate with mature 70S ribosomes.
NusG was purified to 90% homogeneity and tested for activity in an in vitro transcription assay. The purified preparation suppressed pausing and stimulated Rho-dependent termination at the Rho-dependent λtR1 terminator (Supporting Information Fig. S1). Our purified 70S ribosome preparation could synthesize tripeptides using an in vitro translation assay, confirming its activity (Supporting Information Fig. S2, lanes 1–3). To detect association of NusG with ribosomes, we employed size exclusion chromatography (SEC), using a column with a resolution range of 10–600 kDa. We probed for NusG and ribosomes in the various fractions by western blotting with anti-NusG antibody and anti-30S antibody. The latter was made against the S1 protein of the 30S subunit. Since ribosomes are bulky nucleoprotein complexes of about 2.5 MDa and the molecular weight of NusG is 21 kDa, their elution profiles are widely separated by SEC. Therefore, ribosomes were not retained in the column and eluted in the void volume (Fig. 1A, fractions 16–23) whereas purified NusG was trapped and eluted in later fractions (Fig. 1B, fractions 34–37).
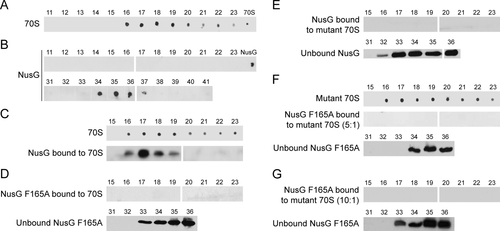
Western blots and western dot blots of Superdex-200 column fractions. (Sections from separate blots were approximated to display all the required fractions. NusG/NusG F165A and ribosome/mutant ribosome preparations were used as controls on all western blots even if not displayed)
A. 70S ribosome elution profile. Fractions 11–23.
B. Wild type NusG elution profile. Fractions 11–23 and 31–41.
C. NusG and 70S (5:1) complex. Fractions 15–23 probed for ribosomes and NusG.
D. NusG F165A and 70S (5:1) combination. Fractions 15–23 and 31–36 probed for NusG.
E. NusG and mutant 70S (5:1) combination. Fractions 15–23 and 31–36 probed for NusG.
F. NusG F165A and mutant 70S (5:1) combination. Fractions 15–23 probed for ribosomes and NusG, fractions 31–36 probed for NusG.
G. NusG F165A and mutant 70S (10:1) combination. Fractions 15–23 and 31–36 probed for NusG.
When we incubated purified NusG and 70S ribosomes in a 5:1 molar ratio for 30 min at room temperature and then applied the mixture to the column, we detected NusG in the void volume, indicating association with ribosomes (Fig. 1C, fractions 16–19). A major fraction of NusG remained unbound and eluted in fractions 34–37 (not shown). These results indicate that NusG and ribosomes can form a complex in vitro, and that no accessory protein or nucleic acid factors are required to promote this association.
Demonstrating NusG-ribosome interaction with a pelleting assay
The above data demonstrated an in vitro interaction between NusG and 70S ribosomes using SEC. To eliminate possible artifacts associated with this technique, we employed an alternative method to detect the interaction. The pelleting assay is used to fractionate proteins based on their density and sedimentation rate in a high-concentration sucrose solution. We therefore exploited differential sedimentation as an alternative method to ask if NusG and ribosomes associate in vitro.
Wild-type NusG and wild-type ribosomes were pre-incubated separately or together at a 5:1 molar ratio, overlaid on a sucrose solution and subjected to ultracentrifugation. The pellets and the supernatants were analyzed by Western blotting. Bulky ribosomes were recovered in the pellet, whereas NusG alone did not pellet under these conditions. NusG was found in the pellet only when NusG and ribosomes were pre-incubated together, confirming their association in vitro (Fig. 2). We measured and compared the pixel counts of the bound NusG in the pellet fraction of NusG + 70S vs total NusG from supernatant and pellet fractions of NusG + 70S in three separate experiments. We observed that 19% (SD = 3) of the total NusG loaded was recovered in the ribosomal pellet fraction as would be expected for a 1:1 NusG:70S association in a 5:1 molar mixture.
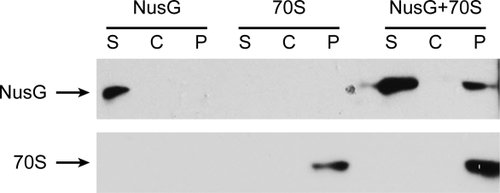
NusG:ribosome association in vitro using pelleting assay. Supernatant (S), sucrose cushion (C) and pellet (P) probed for presence of NusG and ribosomes.
NusG F165A does not associate with wild-type ribosomes
To confirm the specificity of NusG binding to ribosomes, we tested NusG carrying a point mutation in the NusG:S10 binding interface. The F165 residue in the NusG-CTD is highly conserved in bacteria, and the NMR-based structure predicts that mutations in this residue will disrupt the interface (Burmann et al., 2010).
The activity of purified NusG F165A was tested in an in vitro transcription assay. The mutant suppressed pausing to the same extent as wild-type NusG. However, unlike the wild-type protein, NusG F165A inhibited, rather than stimulated, Rho termination at λtR1 (Supporting Information Fig. S1). This is not unexpected, NusG F165A can bind to RNAP (and suppress pausing) but is defective in Rho binding.
Purified NusG F165A was incubated with 70S ribosomes in a 5:1 molar ratio and the mixture analyzed by SEC. In contrast to wild-type NusG, NusG F165A was not detected with ribosomes in the void volume fractions (Fig. 1D, 16–23), indicating that the mutation ablated association of NusG F165A with the ribosome. As expected, unbound NusG F165A was recovered in lower molecular weight fractions (Fig. 1D, 33–36).
Wild-type NusG does not associate with S10 mutant ribosomes
Structural studies revealed S10 residues that lie at the interface with the NusG-CTD (Burmann et al., 2010). Using single-stranded oligonucleotide recombineering, we generated single mutation alanine derivatives of S10 interface residues at M88 and at D97 as well as the double mutant M88/D97A. We were, however, unable to construct single, double or triple mutant derivatives of another interface residue, I100A, which implies that this mutation is lethal in E. coli.
The M88/D97A S10 mutant ribosomes were purified and tested for activity in an in vitro translation assay. The preparation carried out tripeptide synthesis in vitro, indicating that the mutant ribosomes retained protein synthetic activity (Supporting Information Fig. S2, lanes 4–6). However, mutant ribosomes were only ∼70% as active as wild-type ribosomes. Possibly, the mutant ribosome preparation contained more dissociated ribosomes, or the mutant ribosomes were less able to support translation. The S10 mutant ribosomes were then tested for binding with wild-type NusG using the SEC assay. As predicted from the structural studies, wild-type NusG could not bind S10 mutant ribosomes in vitro (Fig. 1E), as indicated by the absence of NusG in the void volume fractions of the column.
NusG F165A does not bind NusE M88/D97A mutant ribosomes
These results with mutant NusG F165A or mutant NusE M88/D97A ribosomes clearly demonstrated that amino acid substitutions in the binding interface between S10 and NusG eliminate NusG:ribosome association. This supports the notion that there is a specific interaction between NusG and ribosomes. To provide further evidence for this idea, we incubated mutant NusG and mutant ribosomes in a 5:1 molar ratio and purified the mixture by SEC. As expected, NusG F165A was not detected in the void volume fractions of the column (Fig. 1F). To screen for a possible weaker interaction, we increased the molar ratio to 10:1. Again, no binding of NusG F165A to mutant ribosomes (NusE M88/D97A) was detected (Fig. 1G).
Evidence for NusG-ribosome association in vivo
Our model for transcription-translation coupling proposes that ribosomes sequester the NusG-CTD in the translational complex, whereas the NusG-NTD is bound to RNAP. Upon encountering stop codons or rare codons, NusG-CTD dissociates from the released ribosomes while remaining bound to RNAP (Roche and Sauer, 1999; Harinarayanan and Gowrishankar, 2003; R. Washburn, unpublished). The unbound NusG-CTD can now bind Rho and stimulate transcription termination. Although there is evidence for in vivo association of Rho and NusG (Li et al., 1993), there is yet no evidence that NusG associates with translating ribosomes in vivo. We therefore attempted to demonstrate this association.
MDS42, MDS42 nusE M88/D97A and MDS42 nusG F165A cells lysed to prepare crude ribosome preparations (see Experimental Procedures) containing a mixture of free and translating ribosomes, were analyzed for NusG content using SDS PAGE electrophoresis and Western blotting.
As shown in Fig. 3, NusG was present in the crude ribosome fraction of wild-type cells. The level of ribosome-bound NusG was significantly reduced in preparations obtained from NusE M88/D97A or NusG F165A mutants. We conclude that NusG and ribosomes associate in vivo via the NusG-CTD and the S10 ribosomal subunit. Mutating the binding interface in either NusG or S10 weakens the interaction between NusG and ribosomes in vivo, rather than abolishing it entirely, as in the in vitro experiments.
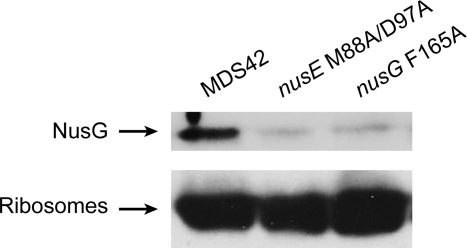
NusG:ribosome association in vivo. Equal quantities of crude ribosome preparations from MDS42, MDS42 nusE M88/D97A (11601) and MDS42 nusG F165A (10780) were probed for NusG (top panel) and ribosomes (lower panel). The slight misalignment of bands was caused by the presence of sucrose in the samples.
Mutations reducing NusG-ribosome interaction inhibit coupling in vivo
Coupling of transcription with translation prevents RNAP backtracking, replisome clashes and formation of lethal DNA double-strand breaks (Dutta et al., 2011). To ask if the NusG and S10 interface mutations lead to uncoupling, we slowed translation elongation by growing cells on agar containing sub-lethal concentrations of chloramphenicol. In the absence of chloramphenicol, neither the nusE M88/D97A nor nusG F165A mutations decreased plating efficiency or colony size after 24 h (data not shown) or 48 h incubation (Fig. 4, LB).
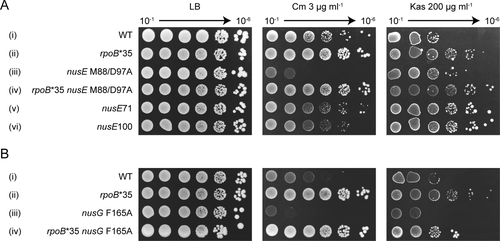
Mutations in the NusG:ribosome interface affect transcription-translation coupling in vivo. Colony-forming ability was assayed in the presence of 3 µg ml−1 chloramphenicol (Cm) targeting translation elongation and 200 µg ml−1 kasugamycin (Kas) targeting translation initiation.
A. The effect of nusE M88/D97A and other S10 mutations on strain growth in the presence of antibiotics. In every strain the nusE gene is linked to tetracycline-resistant Tn10. The strain order from top to bottom: 10401, KM821, 11601, KM820, 10400, KM812.
B. The effect of nusG F165A mutation on strain growth in the presence of antibiotics targeting translation. In all strains the nusG gene is linked to a kanamycin resistance cassette. The order of strains from top to bottom: KM848, KM852, KM850, KM854.
Challenging the nusE M88/D97A mutant with 3 µg ml−1 chloramphenicol significantly inhibited growth (Fig. 4A iii). In contrast, the wild-type nusE strain showed only a minor growth defect (Fig. 4A i). To confirm that this phenotype is the result of transcription-replication clashes, the RNAP mutation, rpoB*35, which suppresses such clashes (Trautinger et al., 2005), was introduced into the nusE M88/D97A mutant. As predicted, rpoB*35 restored the ability of nusE M88/D97A to grow on chloramphenicol. We note that rpoB*35 generally enhances cell growth. This phenotype is, at present, unexplained.
To establish that chloramphenicol sensitivity is specific to mutations in the NusG:S10 interface, we tested the growth of two other nusE mutants, nusE71 (A86D) (Court et al., 1995) and nusE100 (R72G) (Luo et al., 2008). nusE71 is located in the globular domain of S10, far from NusG-binding interface, in a region which possibly interacts with RNAP (Court et al., 1995; Luo et al., 2008). The nusE71 mutant is defective in N-dependent antitermination, and prevents phage λ infection (Friedman et al., 1981). The nusE100 mutation on the other hand abolishes interaction of S10 and NusB and was first characterized as restricting termination by HK022 Nun but not λ N-antitermination (Robledo et al., 1991; Luo et al., 2008). Strains containing nusE71 and nusE100 mutations grew as well as the wild-type parent in the presence of chloramphenicol, although with slightly reduced colony size (Fig. 4A v, vi and i respectively). Therefore, of the nusE mutations tested, only nusE M88/D97A, which reduces NusG binding, induced chloramphenicol sensitivity, presumably by uncoupling transcription and translation.
To confirm that the chloramphenicol sensitivity of nusE M88/D97A stems from impaired interaction of S10 with NusG, we next exposed the nusG F165A mutant to 3 µg ml−1 chloramphenicol. The nusG F165A mutant was 100-fold more sensitive to chloramphenicol compared to the parent strain (Fig. 4B iii and i respectively). This sensitivity was also reversed by rpoB*35 (Fig. 4B ii and iv).
Kasugamycin inhibits translation initiation rather than translation elongation (Woodcock et al., 1991). At 200 μg ml−1, kasugamycin inhibited the growth of the wild-type strain approximately 100-fold (Fig. 4A i). The rpoB*35 mutation suppressed this sensitivity (Fig. 4A ii). In contrast to chloramphenicol, the nusE M88/D97A strain grew better than the WT in the presence of kasugamycin, as did nusE71 and nusE100 (Fig. 4A iii, v and vi). The nusG F165A mutant grew similarly to the wild-type on kasugamycin, and the rpoB*35 mutation enhanced the growth of both wild-type and the nusG mutant (Fig. 4B). Thus, the nusG F165A mutant is not defective in translation initiation.
These results show that strains with weakened NusG:S10 interaction are prone to ribosome uncoupling – and premature transcription termination – when translation elongation is slowed by, for example, chloramphenicol.
Discussion
The 21 kDa EcNusG protein belongs to the Spt5 family, which is characterized by a NusG N-terminal domain (NGN) that interacts with RNAP and a C-terminal KOW domain that binds protein partners. The NusG KOW binds S10/NusE, NusA and Rho transcription termination factor (Werner, 2012; Tomar and Artsimovitch, 2013; Strauß et al., 2016). The E. coli NusG NGN and KOW domains are connected by a flexible linker (Mooney et al., 2009b), which allows NusG to act as a bridging molecule. An early observation that NusG depletion slowed translation strongly implicated NusG in transcription-translation coupling (Zellars and Squires, 1999). A recent study demonstrated that Nus factors support rRNA maturation and ribosome assembly, again pointing to their role in translation (Bubunenko et al., 2013). NusG is one of very few proteins that are conserved across all domains of life (Werner, 2012). In addition to NusG, E. coli carries a NusG paralogue, RfaH, which links transcription with translation in genes with an upstream ops element. These genes lack a consensus ribosome-binding site, and it is thought that RfaH recruits the first ribosome through linkage to the transcription elongation complex (Tomar and Artsimovitch, 2013).
EcNusG and Rho-dependent termination
EcNusG stimulates Rho-dependent transcription termination both in vitro and in vivo. NusG slows the off-rate of Rho from nascent RNA, stimulates Rho-mediated dissociation of transcription complexes (Li et al., 1993; Nehrke and Platt, 1994; Burns et al., 1999). Recent work shows that NusG acts in the central channel of Rho, promoting faster isomerization of the open to the closed hexameric states during its RNA-loading step (Valabhoju et al., 2016). The nusG F165A mutation reduces Rho termination at λtR1 (Supporting Information Fig. S1). Enhancement of Rho activity is the essential function of NusG in E.coli. Therefore, NusG can be ablated in E. coli carrying a rac prophage deletion (Cardinale et al., 2008). Rho-transcription termination is required to prevent expression of the lethal kil gene from the rac prophage pRM promoter. However, although a nusG deletion strain survives, it grows poorly and dies in stationary phase (data not shown).
Unlike NusG, Rho cannot be ablated in E. coli. Rho prevention of lethal R-loop formation (Leela et al., 2013), antisense transcription (Peters et al., 2012) and replisome clashes with stalled transcription elongation complexes (Dutta et al., 2011; Washburn and Gottesman, 2011) presumably account for its essentiality.
Rho-dependent termination and coupling
A model has been proposed to explain the roles of Rho, NusG, RNAP and ribosomes in transcription-translation coupling. The transcription elongation complex is bound to NusG-NTD whereas the lead ribosome is bound to NusG-CTD via the S10 subunit. The interface between NusG-CTD and S10 overlaps with the interface between NusG-CTD and Rho (Burmann et al., 2010). Therefore, coupling prevents NusG-CTD from activating Rho within genes. Ribosome release or stalling uncouples the transcription elongation complex, freeing the NusG-CTD to interact with Rho. When translation is not inhibited, transcription termination occurs only at the ends of genes after ribosome release.
We have determined that NusG can bind to ribosomes in the absence of additional factors. It is not known whether NusG first binds RNAP and recruits ribosomes, binds first to ribosomes, or binds simultaneously to both RNAP and ribosomes. CHiP-chip analysis suggests that NusG binds to RNAP in vivo only after significant RNA synthesis (Mooney et al., 2009a). However, a pause signal located near the transcription start site may allow NusG to couple the lead ribosome to RNAP (Larson et al., 2014; Vvedenskaya et al., 2014). Similarly, RNAP paused at the ops signal is critical for RfaH to couple transcription and translation (Artsimovitch and Landick, 2002). We note that the in vivo calculated numbers for NusG (≈10,000 copies/cell, [Li et al., 1992; Torres et al., 2004]) is one sixth the number of ribosomes (Bremer and Dennis, 2008). This raises the possibility that only the first ribosome in a polysome is associated with NusG.
Three recent papers report structural evidence for direct ribosome to RNAP contacts. Fan et al. (2017) have purified a ribosome and RNAP complex. Although NusG was added to their mixture, it did not appear in the final complex. Kohler et al. (2017) describe a cryo-EM structure based on translation of mRNA up to a stalled TEC in the absence of NusG. The structure of the complex will not permit NusG to span between the ribosome and RNA polymerase. Lastly, Demo et al. (2017) have determined a cryo-EM structure of a 30S subunit and RNAP complex. The Demo and Kohler structures do not agree. In the latter structure, the RNA exit tunnel of RNAP aligns with the Shine-Dalgarno-binding site of the 30S subunit, and importantly, the nucleic-acid-binding cleft of RNAP is detected in distinct conformations. These authors suggest that the transcription-translation complex can assume different functional states during transcription-translation coupling, one of which could include NusG. Our studies do not exclude a NusG-independent coupling pathway, but provide direct evidence for specific NusG binding to 70S ribosomes, which along with known interactions between NusG and RNAP, indicate that NusG can couple transcription with translation in vivo.
Coordination of transcription and translation in eukaryotes
It seems likely that eukaryotes, like bacteria, coordinate transcription and translation. How might this be accomplished when these two processes occur in separate cellular compartments?
We suggest that Spt5 may also coordinate the eukaryotic transcription with translation. Harel-Sharvit et al. (2010) showed that two subunits of RNA polymerase II, Rpb4 and Rpb7, dissociate from PolII and enter the cytoplasm bound to nuclear transcripts. The RNA-bound subunits control translation initiation and mRNA stability. Rpb4/7 are bound to the KOW domain of Spt5 in RNAP II. It is possible that signals from the cytoplasm could act upon Spt5 to modulate this association (Li et al., 2014).
Experimental procedures
Bacterial strains and plasmids
Table 1 shows a list of strains and plasmids used in this study. The list of intermediate strains as well as the description of strains and plasmids construction can be found in Supporting Information data.
| Strain | Genotype | Construction/Reference |
|---|---|---|
| 10400 | MDS42 nusE71:Tn10 | P1.7006 × MDS42 |
| 10401 | MDS42 nusE+:Tn10 | P1.7006 × MDS42 |
| 10780 | MDS42 rpoC:[His6-kan] nusG F165A | P1.NB885 × MDS42 |
| 11601 | MDS42 nusE M88/D97A:Tn10 | P1.SS2132 × MDS42 |
| BLR | BL21 DE3 recA [F- ompT hsdSB( ) gal dcm (DE3) Δ(srl-recA)306::Tn10] ) gal dcm (DE3) Δ(srl-recA)306::Tn10] |
Novagen |
| KM812 | MDS42 nusE100:Tn10 | P1.10289 × MDS42 |
| KM820 | MDS42 rpoB*35 nusE M88/D97A:Tn10 | P1.11601 × 12422 |
| KM821 | MDS42 rpoB*35 nusE+:Tn10 | P1.11601 × 12422 |
| KM848 | MDS42 nusG:<kan> | dsDNA recombineering into MDS42 using pKD46 |
| KM850 | MDS42 nusG F165A:<kan> | dsDNA recombineering into MDS42 using pKD46 |
| KM852 | MDS42 rpoB*35 nusG:<kan> | dsDNA recombineering into 12422 using pKD46 |
| KM854 | MDS42 rpoB*35 nusG F165A:<kan> | dsDNA recombineering into 12422 using pKD46 |
| MDS42 | MG1655 deleted for ∼14% of the genome | (Posfai et al., 2006) |
| MRE600 | rna ΔhdsM | (Cammack and Wade, 1965; Kurylo et al., 2016) |
NusG protein purification
His-tagged NusG and NusG F165A were purified from BLR strain (BL21 DE3 recA, Novagen) transformed with pRM431 or pCu2121 respectively. Cell preculture was raised overnight in LB containing ampicillin (100 μg ml−1) at 37°C, shaking at 235 rpm and sub-cultured 1:100 in LB-Amp at 37°C, until the culture reached A600 = 0.7. The culture was then induced with IPTG, grown for another 3–4 h and harvested 4347 × g for 15 min in a JA25.5 rotor. Supernatant was decanted and the pellet was frozen at −80°C until further use. The cell pellet was thawed from −80°C on ice and dissolved in 10 ml lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, EDTA-free protease inhibitor from Sigma-Aldrich) per gram cell weight. The cells were taken through four freeze-thaw cycles in a dry-ice ethanol bath and water at room temperature and then sonicated for 4 × 15 s pulses. The supernatant was collected by centrifugation at 12,000 × g, 4°C, for 30 min in the JA25.5 rotor. The Ni-NTA resin (GE Healthcare) was washed in lysis buffer and the clear supernatant was added to it (5 ml resin/10 ml lysate) and allowed to bind with rotation overnight at 4°C. The resin was packed in 2 ml columns and washed with five bed-volumes of lysis buffer. Elution was performed with a gradient of 100–500 mM imidazole. Pooled fractions were dialyzed against binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 1 mM TCEP) and purified through a Superdex 200 size exclusion column (GE Life Sciences). Protein was stored in the binding buffer with 50% glycerol in aliquots at −80°C.
Ribosome purification
Both wild type and mutant ribosomes were purified using a protocol developed in the laboratory of J. Frank (Columbia University). Strain MRE600 was used for purification of wild-type ribosomes and 11601 for purification of nusE M88/D97A mutant ribosomes. MRE600 and 11601 strains were shaken in LB overnight at 37°C. The cells were sub-cultured 1:100 in 1 l LB and grown under shaking at 37°C. At A600 = 0.8 the cells were transferred to 4°C to halt growth and harvested by centrifugation at 6600 × g for 20 min at 4°C in a JLA 10.5 rotor. The pellet was suspended in suspension buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl) at 2x the weight of cells and centrifuged at 48,000 × g for 20 min at 4°C in a JA 25.5 rotor. The pellet was stored at −80°C until further use. The cells were then thawed on ice and suspended in 28 ml of opening buffer (20 mM Tris-HCl, pH 7.5, 100 mM NH4Cl, 10.5 mM magnesium acetate, 0.5 mM EDTA, 1 mM of TCEP) with RNase-free DNase, (Roche, final concentration 1 μg ml−1) and protease inhibitor cocktail (Roche, EDTA-free, half tablet). The cells were lysed by passing through a French Press 3 times at 800 psi. Supernatant was collected by centrifugation in a Ti70.1 rotor at 36,600 × g for 40 min at 4°C. The supernatant was applied to 5 ml of sucrose cushion (∼33%: 20 mM Tris-HCl, pH 7.5, 500 mM NH4Cl, 10.5 mM magnesium acetate, 0.5 mM EDTA, 1.1 M sucrose, 1 mM TCEP) and centrifuged in Ti70.1 rotor at 82,500 × g for 17–19 h at 4°C. The pellet was washed in 2 ml of washing buffer (20 mM Tris-HCl, pH 7.5 500 mM NH4Cl, 10.5 mM magnesium acetate, 0.5 mM EDTA, 1 mM TCEP) and then suspended in 1–2 ml of washing buffer. The suspension was applied to a 5 ml sucrose cushion again and centrifuged in a Ti70.1 rotor at 71,800 × g for 17–19 h at 4°C. The supernatant was discarded and the pellet was washed with 2 ml of washing buffer and suspended in 1–2 ml of washing buffer. A gradient of 10% and 35% sucrose (1 mM TCEP) was prepared and the suspension was layered on top of the gradient and centrifuged at 47,000 × g in a SW28 rotor for ∼18 h at 4°C without using the brake. The sucrose gradient was collected from the bottom using a vacuum pump in fractions of 500 µl each. Fractions showing highest A260 measurements were pooled and the volume in the tube was restored with opening buffer for the next centrifugation.
Centrifugation was done at 112,000 × g for 20 h at 4°C in a Ti70.1 rotor. The supernatant was discarded and the pellet suspended in 100–200 µl of 1x Polymix buffer (95 mM KCl, 5 mM NH4Cl, 5 mM magnesium acetate, 0.5 mM CaCl2, 8 mM putrescine and 1 mM spermidine). The A260 was measured for quantification (A260 = 1 is equivalent to 23 pmol 70S, yield ∼12 µM from 6 l of culture). Intact ribosomes were observed on carbon-coated grids under an electron microscope (data not shown). The ribosomes were snap frozen in liquid nitrogen and stored in −80°C.
Crude ribosome preparation for testing in vivo association of NusG and ribosomes
Escherichia coli strains MDS42, 11601 and 10780 were grown in 500 ml of LB at 37°C until A600 = 0.8. Cells were centrifuged at 4300 × g for 15 min at 4°C in a JA25.5 rotor. The supernatant was discarded and the pellet suspended in 2 ml of binding buffer with protease inhibitor and lysed by sonication in 5x 5 min pulses. Centrifugation was done at 24,300 × g, 4°C for 15 min in SA600 rotor and the supernatant collected. A second round of centrifugation at 18,000 × g, 4°C for 5 min was performed to remove remaining cellular debris. A 1.5 ml of 37.7% sucrose cushion prepared in binding buffer was poured into centrifuge tubes (Beckman Coulter Cat. No. 349622). Five hundred microliter of cell lysate was overlaid on top of this sucrose cushion and centrifuged at 98,000 × g at 4°C for 2 h in a Ti100 rotor. The pellet was suspended in 0.2 ml of binding buffer and used for SDS PAGE followed by western blotting.
Size exclusion binding assay
WT NusG or mutant NusG F165A (1 µM) was mixed with wild-type or mutant ribosomes (5 µM) in binding buffer and incubated at RT for 30 min. The solution was then passed through a Superdex 200 HR 10/30 column (GE Healthcare), and 500 µl fractions were collected. The void volume fractions containing ribosomes and the peak fractions containing NusG protein were analyzed by western blotting.
Pelleting assay
A 20% sucrose cushion was prepared in binding buffer (filtered through a 0.22–0.45 µm filter). One hundred microliters of this sucrose cushion was added to each 230 µl centrifuge tube (Beckman Coulter). Fifty microliters of protein solution containing either 2 µM wild type ribosomes or 10 µM wild type NusG protein or both were maintained at RT for 30 min and then overlaid on top of the sucrose cushion. Centrifugation was done at 314,000 × g for 10 min in a TLA100 rotor with de-acceleration set to zero (protocol adapted from Moreno et al. [1998]).
The tubes were removed and the supernatant (50 µl) was separated. The cushion was discarded and the invisible pellet was suspended in 50 µl of sucrose cushion. The samples were analyzed by SDS PAGE and western blotting. Signal intensities of total NusG loaded and NusG bound to 70S were quantified using the Histogram function in Gimp software in three independent experiments.
Western blotting
Sample fractions from the size exclusion column or pelleting assay were mixed with SDS gel loading dye, heat-denatured at 90°C for 1 min and run on 12% SDS PAGE using running buffer (10x: 250 mM Tris, 35 mM SDS and 1.92 M glycine in a final volume of 1 l). Transfer was performed in transfer buffer (25 mM Tris, 1.92 mM Glycine in a final volume of 800 ml, 200 ml of cold methanol was added before use) at 70 V for 1 h at 4°C. Blocking was done in blocking buffer (5% w/v nonfat dry milk dissolved in PBST: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, 2 ml Tween-20, pH adjusted to 7.2 in a final volume of 1 l) for 1 h, shaking. Primary antibodies were used in the following ratios and durations: NusG Ab – 1:10,000 or 1:20,000 dilution for 2 h to overnight, 30S Ab – 1:3000 dilution; overnight, His Ab – 1:1000 dilution; overnight. All hybridizations were performed at 4°C. Washing was done three times for 10 min each in PBST (0.1%) at RT. Secondary Ab was applied at 1:1000 dilution in PBST for 1 h followed by three 10 min washes with PBST. Chemiluminescence was captured on X-ray films using the ECL kit from Roche.
Polyclonal Ab against E. coli NusG raised in rabbit was obtained as a gift from Dr. Ranjan Sen (CDFD, Hyderabad, India). E. coli 70S polyclonal mouse Ab specific for S1 protein in 30S ribosomal subunit was obtained from Developmental studies Hybridoma Bank, catalog No. 373C93A1. Monoclonal His-tag Ab, Sigma-Aldrich, catalog no. H1029, mouse. Mouse and rabbit HRP conjugated secondary Ab were purchased from GE healthcare.
Antibiotic sensitivity testing
Single colonies were inoculated into 5 ml LB broth and grown with shaking at 37°C, overnight. Cultures were serially diluted ten-fold in M9 salts and 5 µl samples were spotted on LB agar plates with or without antibiotics. Chloramphenicol was added to a final concentration 3 µg ml−1 and kasugamycin to 200 µg ml−1. Plates were incubated at 37°C and photographed after 24 h and 48 h. The experiment was performed in triplicate; a representative dataset is shown.
Acknowledgements
This work was supported using NIH grant no. GM037219 to MEG. We acknowledge the help provided by R. Sen (CDFD) and are grateful to him for the NusG antibody. We thank R. Mooney and R. Landick (Dept. of Biochemistry, University of Wisconsin, Madison) for the NusG protein constructs, B. Huang and R. Gonzalez (Chemistry Dept., Columbia University, NY) for their help with the in vitro translation assay, N. Severina (Biochemistry Dept., Columbia University, NY) for technical help with ribosome preparations and L. Lignitto and P. Mauvy for experimental support.
Author contributions
The study was designed by SS and MEG. SS, RW, KKM, DC and MEG acquired, analyzed and interpreted the data. SS, KKM, RW, MEG wrote the manuscript. NC, DC constructed nusE strains. RW performed in vitro transcription assay and KKM antibiotic sensitivity assay.