Journal list menu
Mitogenomic phylogeny resolves Cuneopsis (Bivalvia: Unionidae) as polyphyletic: The description of two new genera and a new species
Xiao-Ping Wu, Yu-Ting Dai, Nan Yin, Feng-Yue Shu, Zhong-Guang Chen, Liang Guo, Chun-Hua Zhou, Shan Ouyang, Xiao-Chen Huang
Zoologica Scripta ½ First published: 24 January 2022
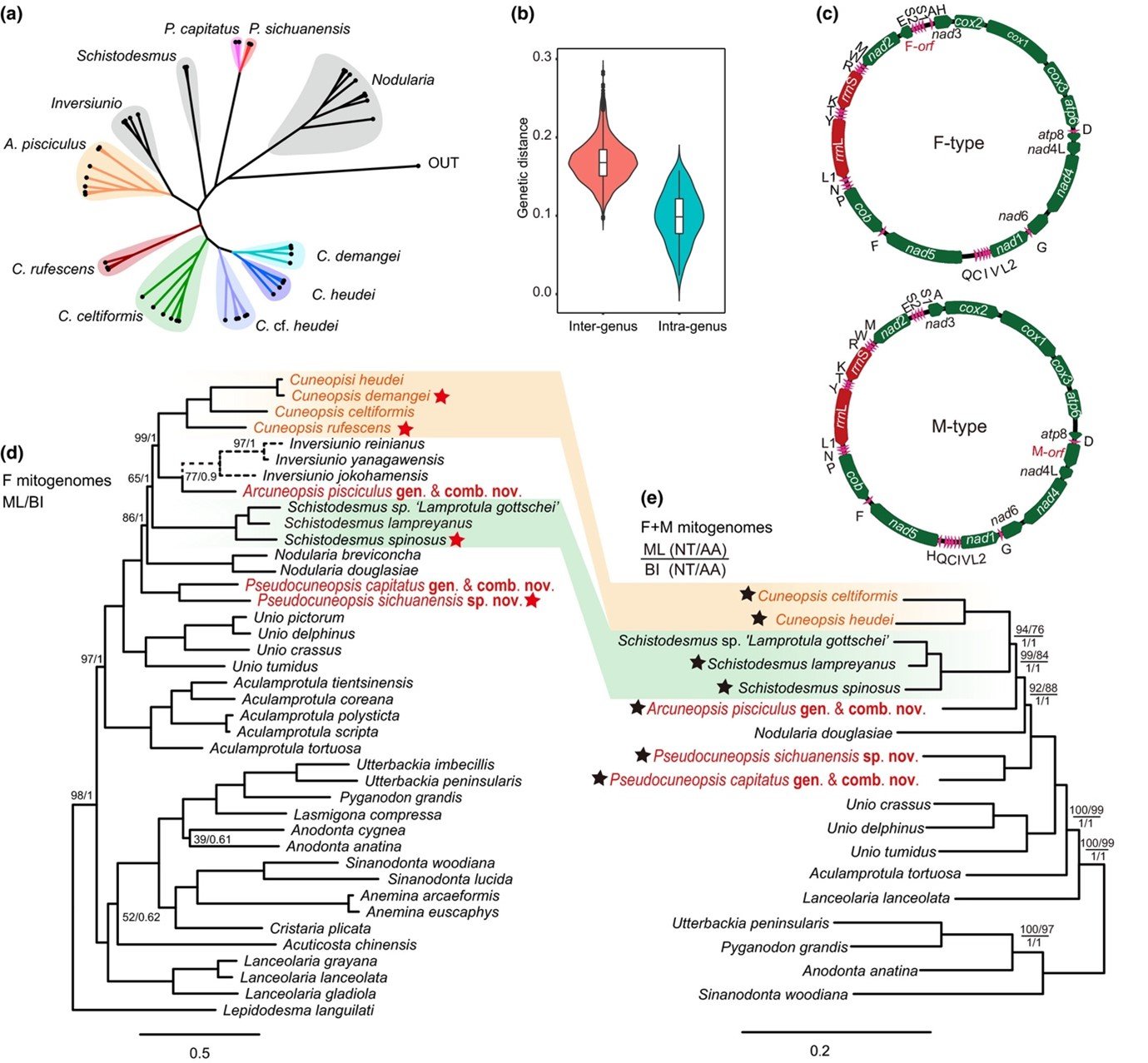
Molecular evidence for the polyphyly of the genus Cuneopsis. (a) Radial cladogram of the maximum likelihood (ML) phylogenetic tree of Cuneopsis and its allied genera inferred from COI sequences. Aculamprotula tientsinensis is used as the outgroup. Each tip marked by a black dot represents a haplotype; (b) Uncorrected COI p-distances in the subfamily Unioninae; (c) Gene maps of the newly generated F- and M-type mitochondrial genomes; (d) Phylogram of the subfamily Unioninae based on Dataset F-NT. The phylogenetic position of Inversiunio (dashed lines) was inferred from partial COI and 16S sequences with other genes treated as missing data; (e) Phylogram of the subfamily Unioninae based on Dataset FM-NT and Dataset FM-AA. ML and BI analyses of mitogenome datasets recovered identical tree topologies. Bootstrap values of ML analysis and Bayesian posterior probabilities are indicated to corresponding nodes, except for those with maximum support. Newly generated F- and M-type mitogenomes are marked by red and black star signs, respectively
Putative ITS2 secondary structure model and multi-gene phylogenies of tetrahymenids (Ciliophora, Hymenostomatia) parasitizing planarians and crayfish worms
Matej Rataj, Peter Vďačný
Zoologica Scripta ½ First published: 15 February 2022
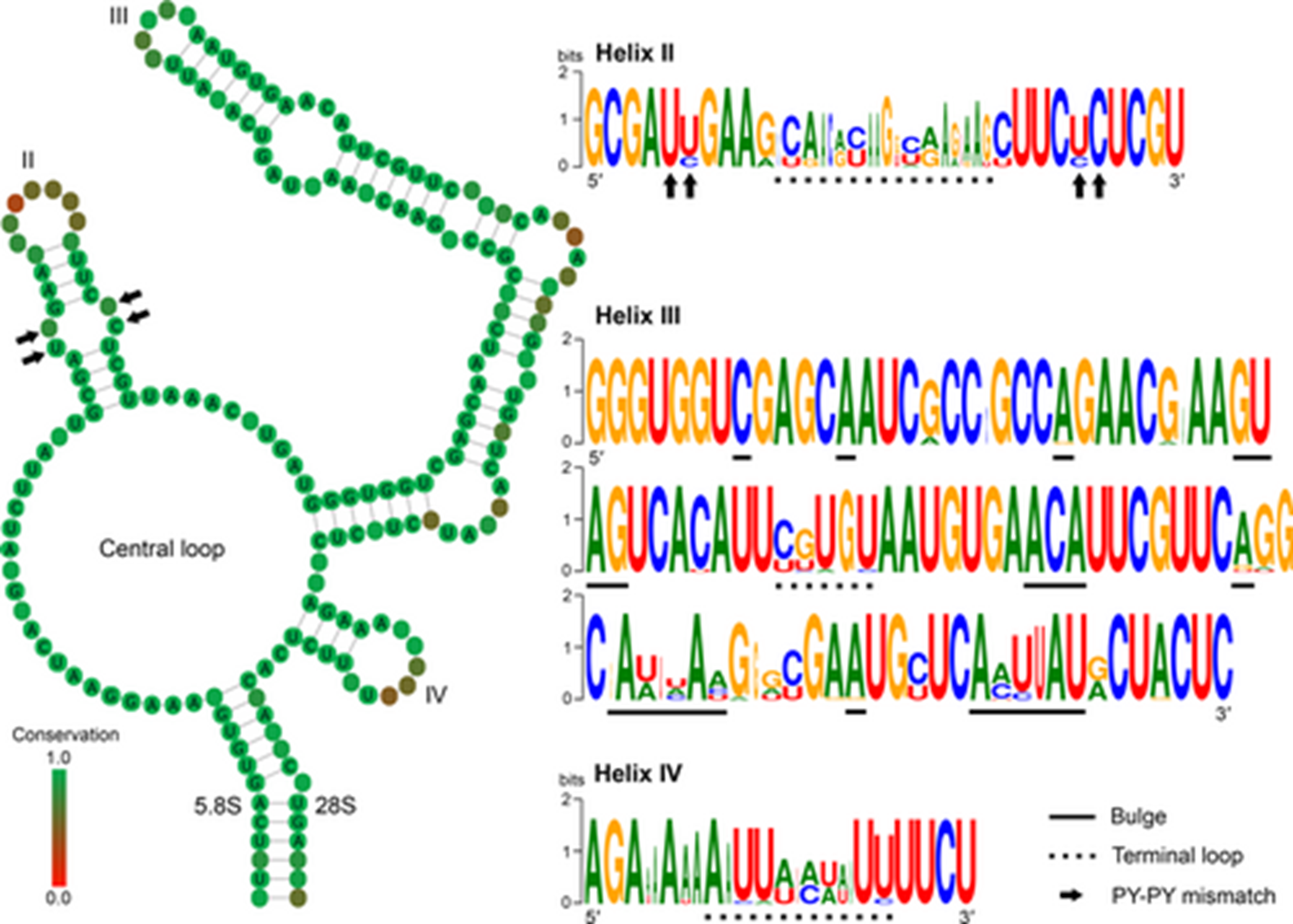
Putative consensus secondary structure of the tetrahymenid ITS2 molecule, showing a central loop with three helices corresponding to helices II, III and IV of other ciliates. The structure logo of helices is shown on the right side. The height of a base is proportional to its frequency in the multiple sequence alignment
Reassessing North Eastern Atlantic-Mediterranean species of Trapania (Mollusca, Nudibranchia)
Sofía Paz-Sedano, José Francisco Martín Álvarez, Terrence M. Gosliner, Marta Pola
Zoologica Scripta ½ First published: 07 April 2022
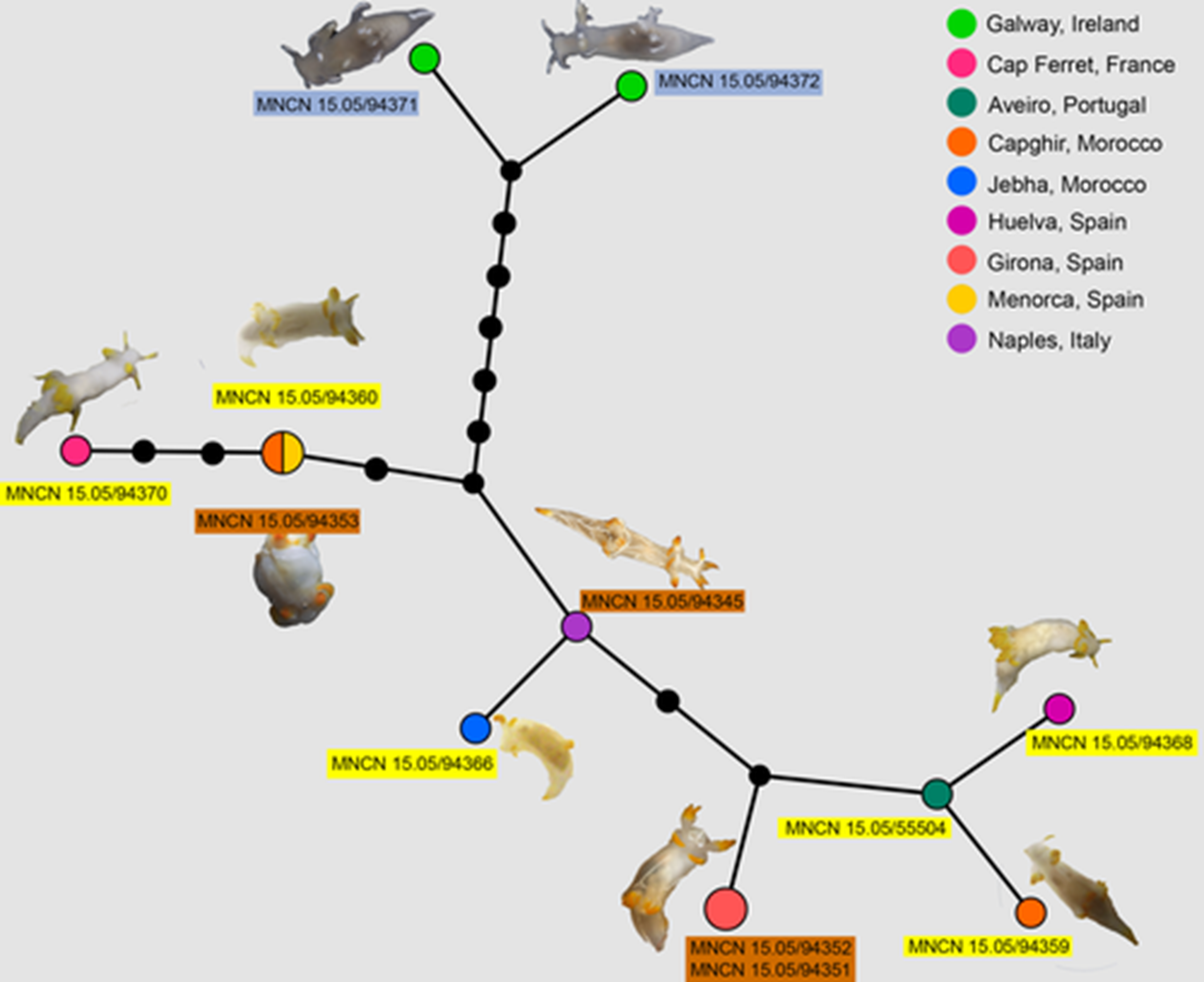
MJ haplotype network analysis based on the COI using PopArt, including sequences of 12 specimens of Trapania lineata. Lines between black dots represent one mutation and black dots represent hypothetical haplotypes. Each coloured circle represents a unique haplotype, and the size of each circle indicates how many specimens share that haplotype. Different colours represent geographic locations. Vouchers within orange, blue and yellow rectangle indicates specimens previously identified as Trapania lineata, Trapania pallida and Trapania hispalensis, respectively.
Neogastropod (Mollusca, Gastropoda) phylogeny: A step forward with mitogenomes
Thomas Lemarcis, Alexander E. Fedosov, Yuri I. Kantor, Jawad Abdelkrim, Paul Zaharias, Nicolas Puillandre
Zoologica Scripta ½ First published: 29 June 2022
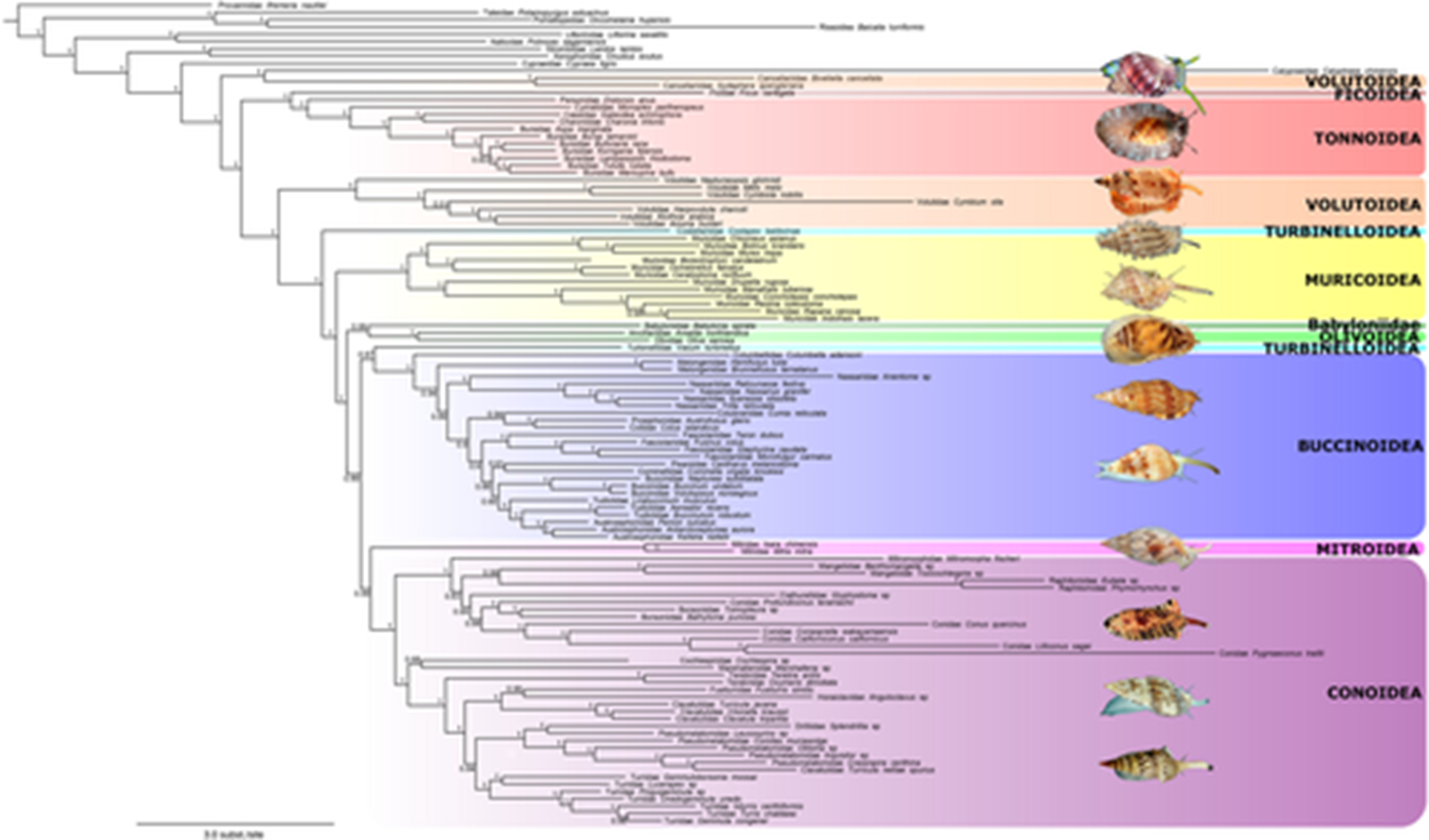
Phylogenetic tree obtained with the NT ‘codon + MFP’ matrix, analysed with MrBayes. The bootstraps values and posterior probabilities (>0.5) are given for each node. Superfamilies are highlighted with colour boxes. Illustrations from top to down: Cancellariidae, Tonnidae, Volutidae, Costellariidae, Muricidae, Olividae, Colubrariidae, Nassariidae, Mitridae, Conidae, Raphitomidae, Turridae (Credits: Philippe Maestrati, Laurent Charles /MNHN). MFP, ModelFinder Plus; NT, nucleotide

Late Cenozoic environmental changes drove the diversification of a weevil genus endemic to the Cape Floristic Region
Noémie M.-C. Hévin, Steffan Hansen, Pia Addison, Laure Benoit, Gael J. Kergoat, Julien Haran
Zoologica Scripta ½ First published: 24 August 2022
Examples of habitat, host plants and habitus in natura for Phlyctinus species. (a) Kogel Bay on the southern coast of the cape floristic region of South Africa with shale fynbos and shale band vegetation types, biotope for several Phlyctinus lineages. (b) Osteospermum moniliferum (top) host of P. ‘sp. n. J2’ (bottom). (c) Dimorphotheca fruticosa (top) host of P. ‘littoralis group 2’ (bottom). (d) Othonna arborescens (top) host of P. ‘sp. n. G' (bottom).
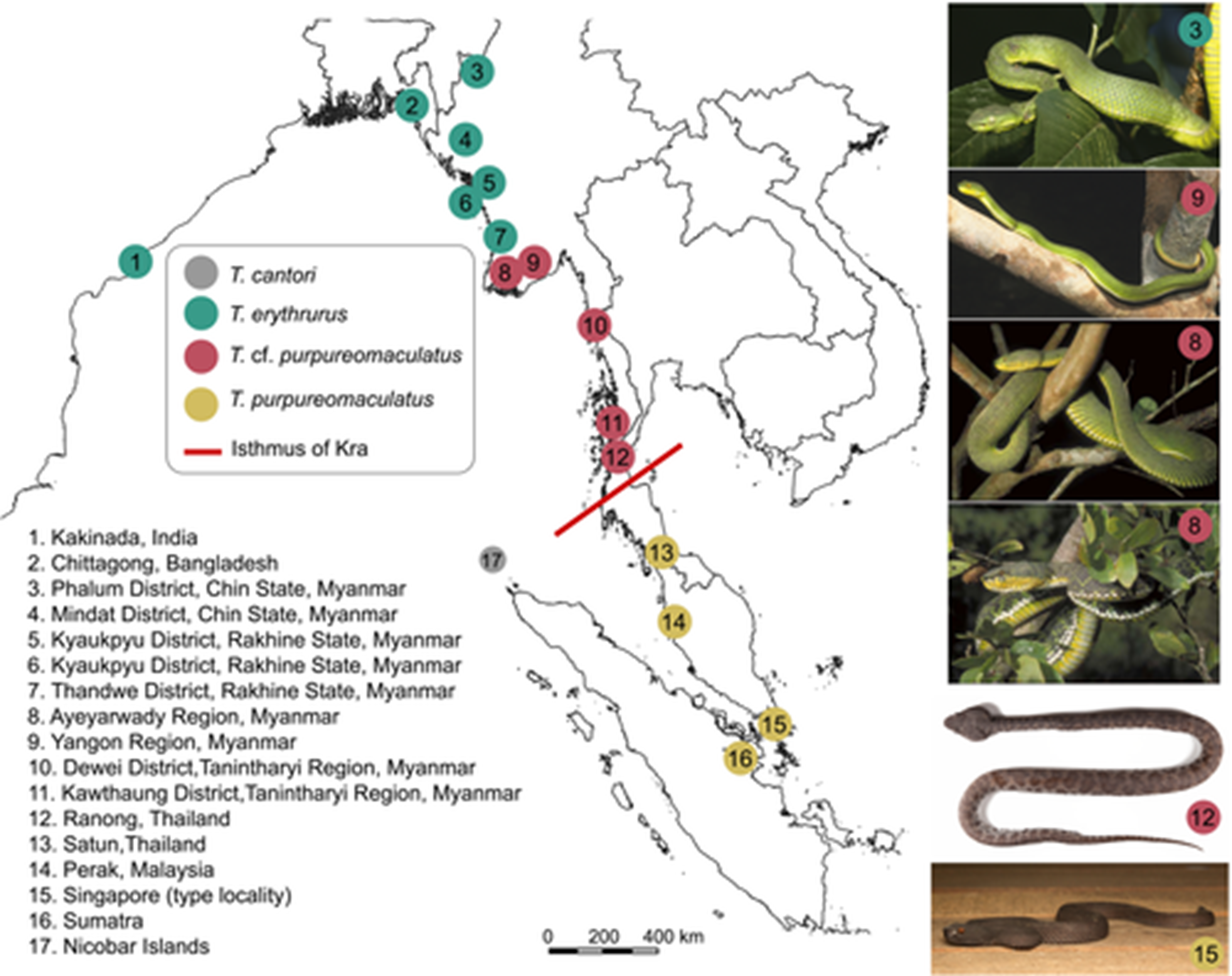
Phylogeography of mangrove pit vipers (Viperidae, Trimeresurus erythrurus-purpureomaculatus complex)
Kin Onn Chan, Law Ing Sind, Law Ingg Thong, Sankar Ananthanarayanan, Shivaram Rasu, Anchalee Aowphol, Attapol Rujirawan, Shahrul Anuar, Daniel Mulcahy, Jesse L. Grismer, L. Lee Grismer
Zoologica Scripta ½ First published: 25 August 2022
Distribution of samples used in this study (left) and their representative colour variants (right). Samples are colour-coded to match phylogenetic clades shown in Figure 3. The museum voucher numbers and specific locality of the representative colour variants from top to bottom are: Trimeresurus erythrurus (CAS235958), Phalum District, Chin State, Myanmar (photo by: Hla tun); T. cf. purpureomaculatus (CAS213410), Hlawga Wildlife Park, Yangon Regi, Myanmar (photo by: CAS-Myanmar Herpetology Survey team, CAS-MHS); T. cf. purpureomaculatus (CAS212245), Mwe Hauk Village, Ayeyarwady Region, Myanmar (photo by: Dong Lin); T. cf. purpureomaculatus (CAS219764), Meinmahla Kyun Wildlife Sanctuary, Pyapone District, Ayeyarwady Region, Myanmar (photo by: Hla tun); T. cf. purpureomaculatus (ZMKU R 00965), Kra Buri District, Ranong Province, Thailand (photo by: AR); T. purpureomaculatus (uncollected specimen), Sungei Buloh Wetland Reserve, Singapore (photo by: LIT). Museum voucher abbreviations are defined in Materials and Methods.
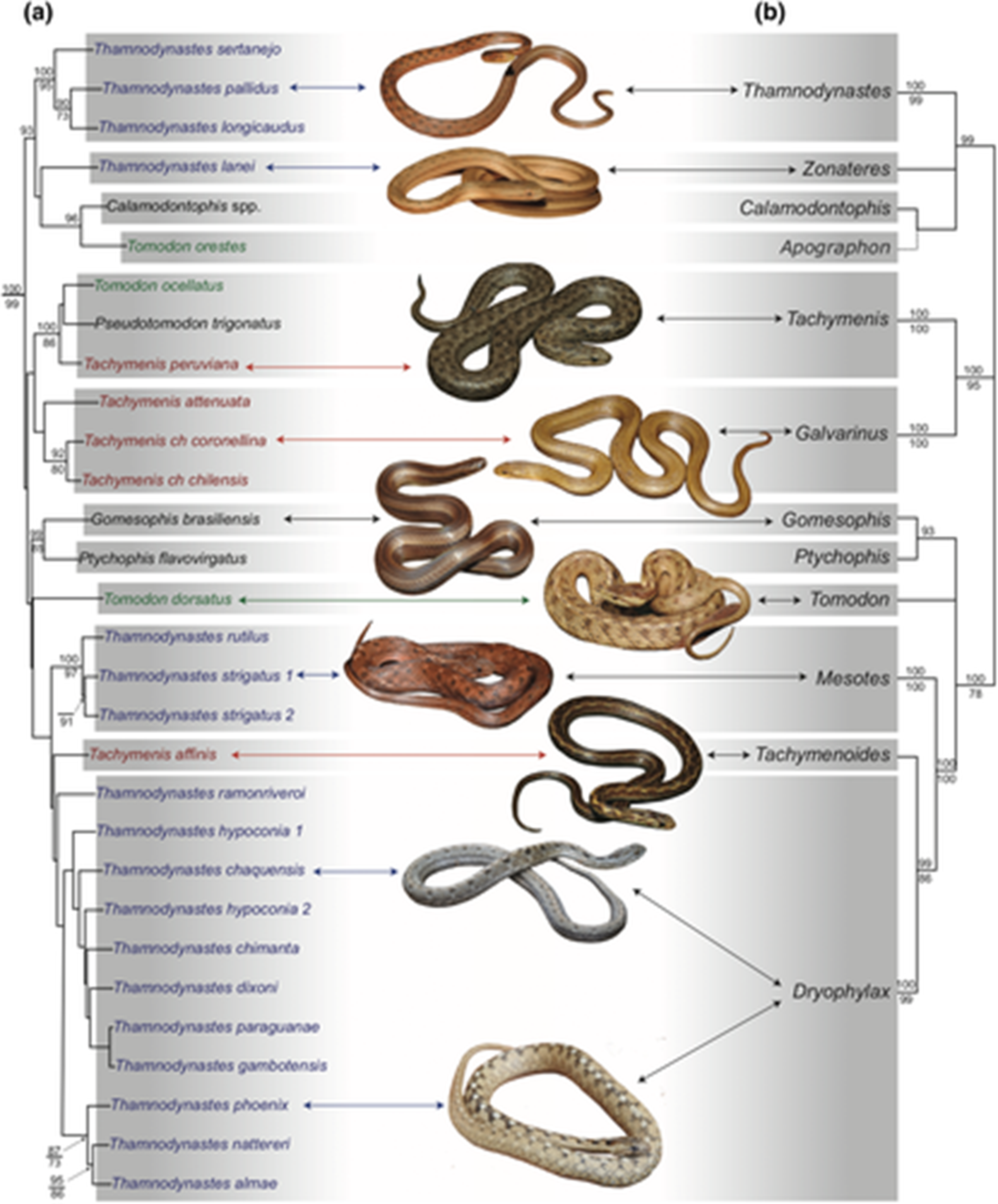
The systematics of Tachymenini (Serpentes, Dipsadidae): An updated classification based on molecular and morphological evidence
Vivian C. Trevine, Felipe G. Grazziotin, Alejandro Giraudo, Nicole Sallaberry-Pincheira, Juliana A. Vianna, Hussam Zaher
Zoologica Scripta ½ First published: 06 September 2022
(a) Total evidence phylogenetic inference of Tachymenini based on the Bayesian topology of morphological and molecular data combined. (b) Concatenated molecular Bayesian topology, collapsed to show the new genus classification. Posterior probability and bootstrap values are indicated above and below branches, respectively. Probability values below 80 and bootstrap values below 70 are not shown. Posterior probabilities clades below 70 are collapsed in (b). Terminals are restricted for Tachymenini, and tip labels are coloured to show no monophyletic genera. The complete topology is provided as (Figure S1). Photo credits: Arthur Abegg (Thamnodynastes pallidus, Gomesophis brasiliensis, Tomodon dorsatus, Thamnodynastes strigatus, and Thamnodynastes phoenix), Boris Blotto (Tachymenis chilensis), Juan Camilo Arredondo (Th. lanei, Tachymenis peruviana, and Tachymenis affinis), and Marcelo Ribeiro Duarte (Thamnodynastes chaquensis)
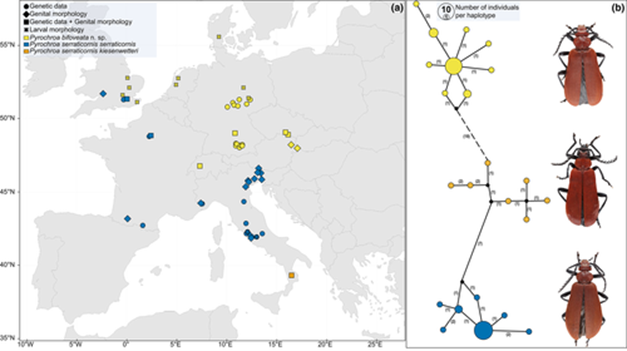
Phylogeny of European Pyrochroa (Coleoptera, Pyrochroidae) reveals cryptic taxa and different glacial histories
Marco Molfini, Emiliano Mancini, Marco A. Bologna
Zoologica Scripta ½ First published: 20 September 2022
(a) Sampling sites distribution map of Pyrochroa bifoveata n. sp. and of the two subspecies of P. serraticornis. Sites for each taxon are marked with a different colour; the dimension of the marker is not proportional to the number of individuals per site. Larval records refer to scientific literature (Graser, 1990; Larsson, 1945; Molfini et al., 2021; van Emden, 1943; van Rossem, 1947). (b) TCS network showing relationship among COI (cytochrome oxidase subunit I) haplotypes from P. bifoveata, P. s. serraticornis and P. s. kiesenwetteri. Sizes of circles are proportional to haplotype frequencies; numbers in parenthesis indicate evolutionary mutational steps; the dashed line divides the two species. A picture of the adult male habitus for each taxon is provided
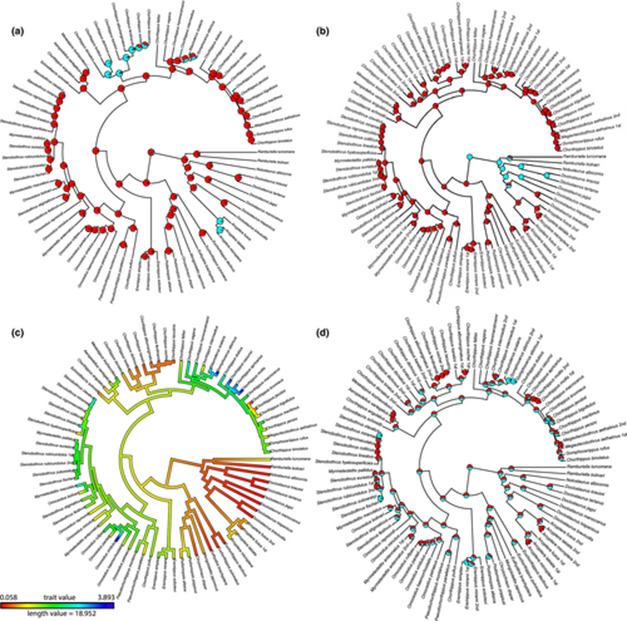
Evolution of calling songs in the grasshopper subfamily Gomphocerinae (Orthoptera, Acrididae)
Nikita Sevastianov, Tatiana Neretina, Varvara Vedenina
Zoologica Scripta ½ First published: 03 January 2023
(a–d) Ancestral state reconstruction of the characters. The pie charts in the nodes show probability of the different character state for a, b and d. (a) character 2, syllable homogeneity (red colour indicates the presence of one syllable type, blue colour – the presence of two syllable types). (b) character 19, usage of two legs (red colour indicates the usage of both legs, blue colour – the usage of only one leg). (c) character 4, echeme duration (colours of the nodes show the most probable value for the character, that is ancestral state estimate; the legend is shown at logarithmic scale). (d) character 13, stepwise downstroke (red colour indicates the absence of stepwise leg-movements, blue colour – the presence of stepwise leg-movements).
The wealth of shared resources: Improving molecular taxonomy using eDNA and public databases
James F. Fleming
Zoologica Scripta ½ First published: 21 February 2023
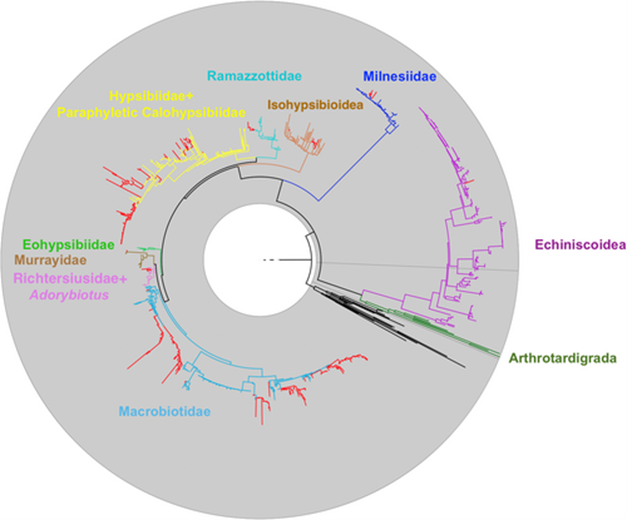
An 18S phylogeny of the Tardigrada, including all 630 sequences previously identified as only “tardigrade 18S” in GenBank. These 18S sequences are identified as red branches, and each family is independently coloured with the outgroup and interstitial branches in black.
Integrative approach resolves the systematics of barred wolf snakes in the Lycodon striatuscomplex (Reptilia, Colubridae)
Amarasinghe A. Thasun Amarasinghe, Rafaqat Masroor, Hmar T. Lalremsanga, Sanjaya Weerakkody, Natalia B. Ananjeva, Patrick D. Campbell, Stevie R. Kennedy-Gold, Sanjaya K. Bandara, Andrey M. Bragin, Atthanagoda K. A. Gayan, Vivek R. Sharma, Amit Sayyed, Lal Biakzuala, Andradige S. Kanishka, Sumaithangi R. Ganesh, Ivan Ineich, Anslem de Silva, Lakshman J. M. Wickramasinghe, Sampath S. Seneviratne, Nikolay A. Poyarkov, Gernot Vogel, Daniel Jablonski
Zoologica Scripta ½ First published: 06 March 2023
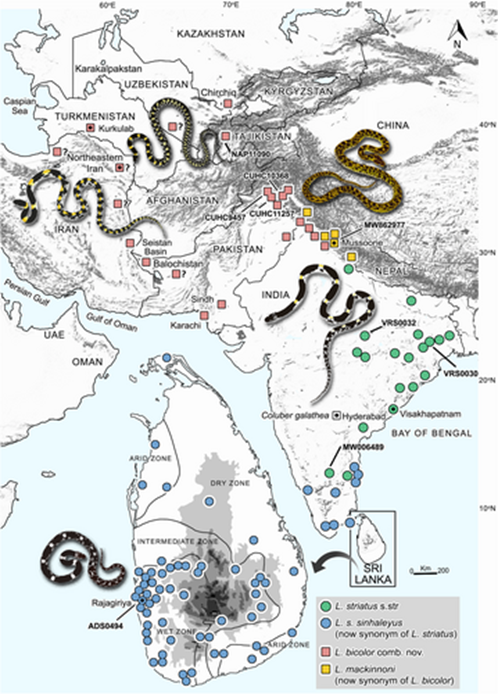
Current distribution showing the museum collection/personal observation/photographic evidence localities of Lycodon striatus sensu stricto in blue and green circles; and L. bicolor comb. nov. in red and yellow squares. The type locality of C. galathea (L. striatus) shows in an open square. The localities of L. mackinnoni (now a junior synonym of L. bicolor comb. nov.) are marked with yellow squares. The symbols with dots in the middle represent the type locality of the respective species. Imprecise localities retrieved from historical publications are marked with a ‘?’ symbol. The localities of genetic samples are marked with GenBank accession numbers.
Mitogenomic phylogeny of Muricidae (Gastropoda: Neogastropoda)
Yi Yu, Lingfeng Kong, Qi Li
Zoologica Scripta ½ First published: 24 April 2023
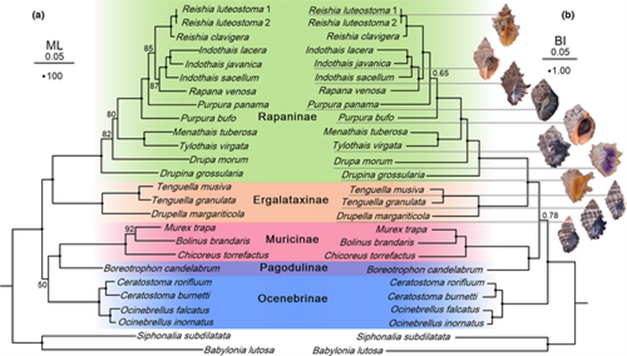
Phylogenetic relationships of Muricidae based on the MA dataset. The ML phylograms are shown in the (a). The BI phylograms are shown in the (b). Numbers at nodes are statistical support values for ML (bootstrap proportions in percentage)/BI (posterior probabilities). Solid black circles represent nodes with posterior probabilities ≥0.95 and bootstrap proportions ≥90. Shells of all the specimens sequenced in this study are illustrated.
Molecular phylogeny of Cercopidae (Hemiptera, Cercopoidea)
Elorde S. Crispolon, Adeline Soulier-Perkins, Eric Guilbert
Zoologica Scripta ½ First published: 02 May 2023
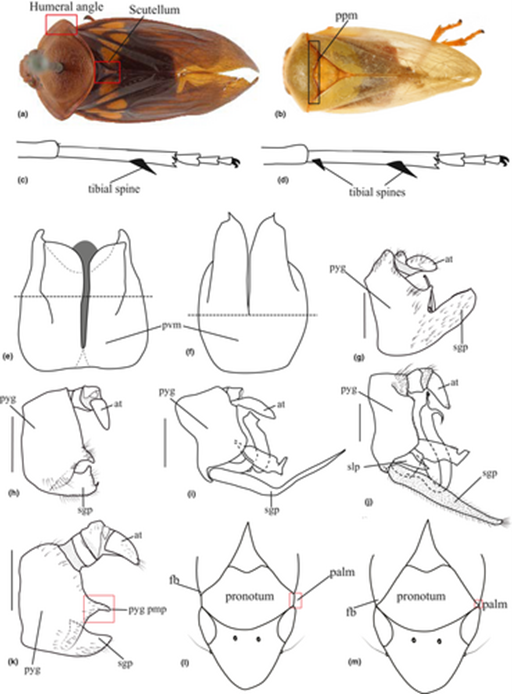
Morphological characters supporting major lineages. (a,b) Habitus in dorsal view showing the humeral angle, scutellum and pronotum posterior margin (a) Suracarta tricolour & (Le Peletier de Saint-Fargeau & Serville, 1825). (b) Trigonoschema negrosensis Crispolon & Yap, 2021. Schematic illustration of: (c) single posterior leg spine and (d) two posterior leg spines. (e,f) Schematic illustration of the pygofer in ventral view. (g–k). Male terminalia in lateral view. (g) Prosapia simulans (Walker, 1858). (h) Cosmoscarta herossa Jacobi, 1921. (i) Aufidus albonigrus Le Cesne & Soulier Perkins, 2021. (j) Wawi mehi Soulier-Perkins and Le Cesne, 2016. (k) Radioscartasp. Schematic illustration of: (l) eye not overlapping the forewing base and not concealing the pronotum antero-lateral margin and (m). eye overlapping the forewing base and concealing the pronotum anter-lateral margin. palm, pronotum antero-lateral margin; ppm, pronotum posterior margin; fb, forewing base; pyg, pygofer; pvm, pygofer ventral margin; at, anal tube; sgp, subgenital plate; pgp pmp, pygofer posterior margin process; slp, sterno-lateral plate.





