The Double-Histidine Cu2+-Binding Motif: A Highly Rigid, Site-Specific Spin Probe for Electron Spin Resonance Distance Measurements†
Timothy F. Cunningham
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorMiriam R. Putterman
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorAstha Desai
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorCorresponding Author
Prof. W. Seth Horne
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)Search for more papers by this authorCorresponding Author
Prof. Sunil Saxena
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)Search for more papers by this authorTimothy F. Cunningham
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorMiriam R. Putterman
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorAstha Desai
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Search for more papers by this authorCorresponding Author
Prof. W. Seth Horne
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)Search for more papers by this authorCorresponding Author
Prof. Sunil Saxena
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)
Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260 (USA)Search for more papers by this authorThis research was supported by a grant from the National Science Foundation (MCB-1157712 to S.S.), and the Bruker E680 instrument used was purchased with funds from the National Institutes of Health (grant 1S10RR028701).
Graphical Abstract
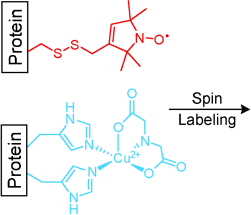
That's great, DEER! When a double-histidine Cu2+-binding motif (shown in blue) assembled in situ from natural amino acid residues and a metal salt was used as a rigid spin probe for double electron–electron resonance (DEER) distance measurements, dramatically narrower and readily interpretable distance distributions were observed than with a commonly used flexible spin label (red). Molecular modeling of an unlabeled protein gave a distance within 0.5 Å of the experimental value.
Abstract
The development of ESR methods that measure long-range distance distributions has advanced biophysical research. However, the spin labels commonly employed are highly flexible, which leads to ambiguity in relating ESR measurements to protein-backbone structure. Herein we present the double-histidine (dHis) Cu2+-binding motif as a rigid spin probe for double electron–electron resonance (DEER) distance measurements. The spin label is assembled in situ from natural amino acid residues and a metal salt, requires no postexpression synthetic modification, and provides distance distributions that are dramatically narrower than those found with the commonly used protein spin label. Simple molecular modeling based on an X-ray crystal structure of an unlabeled protein led to a predicted most probable distance within 0.5 Å of the experimental value. Cu2+ DEER with the dHis motif shows great promise for the resolution of precise, unambiguous distance constraints that relate directly to protein-backbone structure and flexibility.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie_201501968_sm_miscellaneous_information.pdf774.2 KB | miscellaneous_information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1
- 1aW. Hubbell, C. López, C. Altenbach, Z. Yang, Curr. Opin. Struct. Biol. 2013, 23, 725–733;
- 1bG. Jeschke, Annu. Rev. Phys. Chem. 2012, 63, 419–446.
- 2G. Jeschke, Prog. Nucl. Magn. Reson. Spectrosc. 2013, 72, 42–60.
- 3
- 3aR. Langen, K. J. Oh, D. Cascio, W. L. Hubbell, Biochemistry 2000, 39, 8396–8405;
- 3bM. R. Fleissner, D. Cascio, W. L. Hubbell, Protein Sci. 2009, 18, 893–908;
- 3cB. M. Kroncke, P. S. Horanyi, L. Columbus, Biochemistry 2010, 49, 10045–10060;
- 3dD. M. Freed, A. K. Khan, P. S. Horanyi, D. S. Cafiso, Biochemistry 2011, 50, 8792–8803;
- 3eT. F. Cunningham, M. S. McGoff, I. Sengupta, C. P. Jaroniec, W. S. Horne, S. Saxena, Biochemistry 2012, 51, 6350–6359.
- 4
- 4aM. I. Fajer, H. Li, W. Yang, P. G. Fajer, J. Am. Chem. Soc. 2007, 129, 13840–13846;
- 4bY. Polyhach, E. Bordignon, G. Jeschke, Phys. Chem. Chem. Phys. 2011, 13, 2356–2366;
- 4cM. M. Hatmal, Y. Li, B. G. Hegde, P. B. Hegde, C. C. Jao, R. Langen, I. S. Haworth, Biopolymers 2012, 97, 35–44;
- 4dJ. Sarver, J. Townsend, G. Rajapakse, L. Jen-Jacobson, S. Saxena, J. Phys. Chem. B 2012, 116, 4024–4033.
- 5
- 5aL. Columbus, T. Kalai, J. Jeko, K. Hideg, W. L. Hubbell, Biochemistry 2001, 40, 3828–3846;
- 5bN. L. Fawzi, M. R. Fleissner, N. J. Anthis, T. Kalai, K. Hideg, W. L. Hubbell, G. M. Clore, J. Biomol. NMR 2011, 51, 105–114.
- 6D. Toledo Warshaviak, V. V. Khramtsov, D. Cascio, C. Altenbach, W. L. Hubbell, J. Magn. Reson. 2013, 232, 53–61.
- 7M. R. Fleissner, M. D. Bridges, E. K. Brooks, D. Cascio, T. Kalai, K. Hideg, W. L. Hubbell, Proc. Natl. Acad. Sci. USA 2011, 108, 16241–16246.
- 8
- 8aE. Narr, A. Godt, G. Jeschke, Angew. Chem. Int. Ed. 2002, 41, 3907–3910;
10.1002/1521-3773(20021018)41:20<3907::AID-ANIE3907>3.0.CO;2-T CAS PubMed Web of Science® Google ScholarAngew. Chem. 2002, 114, 4063–4066;
- 8bJ. S. Becker, S. Saxena, Chem. Phys. Lett. 2005, 414, 248–252;
- 8cZ. Yang, J. Becker, S. Saxena, J. Magn. Reson. 2007, 188, 337–343;
- 8dB. E. Bode, J. Plackmeyer, T. F. Prisner, O. Schiemann, J. Phys. Chem. A 2008, 112, 5064–5073;
- 8eZ. Yang, M. Ji, S. Saxena, Appl. Magn. Reson. 2010, 39, 487–500.
- 9
- 9aI. M. van Amsterdam, M. Ubbink, G. W. Canters, M. Huber, Angew. Chem. Int. Ed. 2003, 42, 62–64; Angew. Chem. 2003, 115, 64–67;
- 9bG. E. Merz, P. P. Borbat, A. J. Pratt, E. D. Getzoff, J. H. Freed, B. R. Crane, Biophys. J. 2014, 107, 1669–1674;
- 9cJ. H. van Wonderen, D. N. Kostrz, C. Dennison, F. MacMillan, Angew. Chem. Int. Ed. 2013, 52, 1990–1993; Angew. Chem. 2013, 125, 2044–2047.
- 10
- 10aD. Goldfarb in Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences (Eds.: C. R. Timmel, J. R. Harmer), Berlin, Springer, 2012, pp. 163–204;
- 10bM. Ji, S. Ruthstein, S. Saxena, Acc. Chem. Res. 2014, 47, 688–695;
- 10cD. Goldfarb, Phys. Chem. Chem. Phys. 2014, 16, 9685–9699.
- 11
- 11aZ. Yang, M. R. Kurpiewski, M. Ji, J. E. Townsend, P. Mehta, L. Jen-Jacobson, S. Saxena, Proc. Natl. Acad. Sci. USA 2012, 109, E 993–1000;
- 11bZ. Yang, G. Jiménez-Osés, C. J. López, M. D. Bridges, K. N. Houk, W. L. Hubbell, J. Am. Chem. Soc. 2014, 136, 15356–15365.
- 12T. F. Cunningham, M. D. Shannon, M. R. Putterman, R. Arachchige, I. Sengupta, M. Gao, C. P. Jaroniec, S. Saxena, J. Phys. Chem. B 2014, 119, 2839–2843.
- 13
- 13aF. H. Arnold, B. L. Haymore, Science 1991, 252, 1796–1797;
- 13bL. Regan, Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 257–287.
- 14
- 14aR. J. Todd, M. E. Van Dam, D. Casimiro, B. L. Haymore, F. H. Arnold, Proteins Struct. Funct. Genet. 1991, 10, 156–161;
- 14bJ. N. Higaki, R. J. Fletterick, C. S. Craik, Trends Biochem. Sci. 1992, 17, 100–104;
- 14cJ. Voss, L. Salwinski, H. R. Kaback, W. L. Hubbell, Proc. Natl. Acad. Sci. USA 1995, 92, 12295–12299;
- 14dK. Jung, J. Voss, M. He, W. L. Hubbell, H. R. Kaback, Biochemistry 1995, 34, 6272–6277;
- 14eA. J. Nicoll, D. J. Miller, K. Futterer, R. Ravelli, R. K. Allemann, J. Am. Chem. Soc. 2006, 128, 9187–9193.
- 15P. S. Nadaud, I. Sengupta, J. J. Helmus, C. P. Jaroniec, J. Biomol. NMR 2011, 51, 293–302.
- 16M. P. Brandi-Blanco, M. M. de Benavides-Giménez, J. M. González-Pérez, D. Choquesillo-Lazarte, Acta Crystallogr. Sect. E 2007, 63, m 1678–m1679.
- 17J. Peisach, W. E. Blumberg, Arch. Biochem. Biophys. 1974, 165, 691–708.
- 18W. B. Mims, J. Peisach, J. Chem. Phys. 1978, 69, 4921–4930.
- 19
- 19aD. Goldfarb, J. Fauth, Y. Tor, A. Shanzer, J. Am. Chem. Soc. 1991, 113, 1941–1948;
- 19bJ. Hernández-Guzmán, L. Sun, A. K. Mehta, J. Dong, D. G. Lynn, K. Warncke, ChemBioChem 2013, 14, 1762–1771.
- 20
- 20aJ. McCracken, S. Pember, S. J. Benkovic, J. Am. Chem. Soc. 1988, 110, 1069;
- 20bK. I. Silva, B. C. Michael, S. J. Geib, S. Saxena, J. Phys. Chem. B 2014, 118, 8935–8944.
- 21G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C. R. Timmel, D. Hilger, H. Jung, Appl. Magn. Reson. 2006, 30, 473–498.
- 22
- 22aG. Jeschke, A. Bender, T. Schweikardt, G. Panek, H. Decker, H. Paulsen, J. Biol. Chem. 2005, 280, 18623–18630;
- 22bM. G. Finiguerra, M. Prudencio, M. Ubbink, M. Huber, Magn. Reson. Chem. 2008, 46, 1096–1101;
- 22cM. A. Swanson, V. Kathirvelu, T. Majtan, F. E. Frerman, G. R. Eaton, S. S. Eaton, Protein Sci. 2011, 20, 610–620;
- 22dD. Klose, J. P. Klare, D. Grohmann, C. W. Kay, F. Werner, H. J. Steinhoff, PLoS One 2012, 7, e 39492;
- 22eG. Hagelueken, R. Ward, J. H. Naismith, O. Schiemann, Appl. Magn. Reson. 2012, 42, 377–391;
- 22fB. Roux, S. M. Islam, J. Phys. Chem. B 2013, 117, 4733–4739.
- 23
- 23aE. Vanea, C. Gruian, C. Rickert, H. J. Steinhoff, V. Simon, Biomacromolecules 2013, 14, 2582–2592;
- 23bI. D. Sahu, R. M. McCarrick, K. R. Troxel, R. Zhang, H. J. Smith, M. M. Dunagan, M. S. Swartz, P. V. Rajan, B. M. Kroncke, C. R. Sanders, G. A. Lorigan, Biochemistry 2013, 52, 6627–6632;
- 23cA. Bowman, C. M. Hammond, A. Stirling, R. Ward, W. Shang, H. El-Mkami, D. A. Robinson, D. I. Svergun, D. G. Norman, T. Owen-Hughes, Nucleic Acids Res. 2014, 42, 6038–6051;
- 23dJ. Vendome, K. Felsovalyi, H. Song, Z. Yang, X. Jin, J. Brasch, O. J. Harrison, G. Ahlsen, F. Bahna, A. Kaczynska, P. S. Katsamba, D. Edmond, W. L. Hubbell, L. Shapiro, B. Honig, Proc. Natl. Acad. Sci. USA 2014, 111, E 4175–4184;
- 23eI. D. Sahu, B. M. Kroncke, R. Zhang, M. M. Dunagan, H. J. Smith, A. Craig, R. M. McCarrick, C. R. Sanders, G. A. Lorigan, Biochemistry 2014, 53, 6392–6401.
- 24I. Sengupta, P. S. Nadaud, J. J. Helmus, C. D. Schwieters, C. P. Jaroniec, Nat. Chem. 2012, 4, 410–417.
- 25
- 25aS. Y. Park, P. P. Borbat, G. Gonzalez-Bonet, J. Bhatnagar, A. M. Pollard, J. H. Freed, A. M. Bilwes, B. R. Crane, Nat. Struct. Mol. Biol. 2006, 13, 400–407;
- 25bO. Duss, M. Yulikov, G. Jeschke, F. H. Allain, Nat. Commun. 2014, 5, 3669;
- 25cO. Duss, E. Michel, M. Yulikov, M. Schubert, G. Jeschke, F. H. Allain, Nature 2014, 509, 588–592.
- 26M. Qi, A. Gross, G. Jeschke, A. Godt, M. Drescher, J. Am. Chem. Soc. 2014, 136, 15366–15378.
- 27D. Abdullin, N. Florin, G. Hagelueken, O. Schiemann, Angew. Chem. Int. Ed. 2015, 54, 1827–1831; Angew. Chem. 2015, 127, 1847–1851.