Identity and functional characterisation of the transporter supporting the Na+-dependent high-affinity NO3− uptake in Zostera marina L.
Abstract
Zostera marina is a seagrass, a group of angiosperms that evolved from land to live submerged in seawater, an environment of high salinity, alkaline pH and usually very low NO3−. In 2000, we reported the first physiological evidence for the Na+-dependent high-affinity NO3− uptake in this plant. Now, to determine the molecular identity of this process, we searched for NO3− transporters common to other vascular plants encoded in Z. marina's genome. We cloned two candidates, ZosmaNPF6.3 and ZosmaNRT2 with its partner protein ZosmaNAR2. ZosmaNAR2 expression levels increase up to 4.5-fold in Z. marina leaves under NO3−-deficiency, while ZosmaNRT2 and ZosmaNPF6.3 expressions were low and unaffected by NO3−. NO3− transport capacity, kinetic properties and H+ or Na+-dependence were examined by heterologous expression in the Hansenula polymorpha high-affinity NO3− transporter gene disrupted strain (∆ynt1). ZosmaNPF6.3 functions as a H+-dependent NO3− transporter, without functionality at alkaline pH and apparent dual kinetics (KM = 11.1 µM at NO3− concentrations below 50 µM). ZosmaNRT2 transports NO3− in a H+-independent but Na+-dependent manner (KM = 1 mM Na+), with low NO3− affinity (KM = 30 µM). When ZosmaNRT2 and ZosmaNAR2 are co-expressed, a Na+-dependent high-affinity NO3− transport occurs (KM = 5.7 µM NO3−), mimicking the in vivo value. These results are discussed in the physiological context, providing evidence that ZosmaNRT2 is a Na+-dependent high-affinity NO3− transporter, the first of its kind to be functionally characterised in a vascular plant, that requires ZosmaNAR2 to achieve the necessary high-affinity for nitrate uptake from seawater.
1 INTRODUCTION
Plants have evolved various transport systems and integrate complex signals that regulate nitrate uptake in relation to the nitrate concentration in the environment, which may vary up to several orders of magnitude (Tsay et al., 2007; Wang et al., 2018). These nitrate concentrations can be persistently low, below 1 µM, in the ultraoligotrophic waters of the Mediterranean (Pasqueron de Frommeurvault et al., 2016). Therefore, having a high-affinity nitrate uptake system is crucial for the survival of marine autotrophs that depend on nitrate as N-source. The current model proposes that terrestrial vascular plants actively take up nitrate from the medium through proton/nitrate-coupled transport systems, including NITRATE TRANSPORTER 1 (NRT1)/PEPTIDE TRANSPORTER (PTR) family (NPF) and NITRATE TRANSPORTER 2 (NRT2), with different kinetic properties (reviewed in Wang et al., 2018). However, these proton-driven nitrate uptake mechanisms do not fit the physiological evidence for Na+-dependent high-affinity NO3− uptake mechanisms found in seagrasses (Rubio & Fernández, 2019). Our group was the first to describe that mechanism (KM = 2.3 µM NO3−) operating in the mesophyll leaf cells of the seagrass Zostera marina (García-Sánchez et al., 2000). This mechanism was also found in Z. marina root cells, for the high-affinity phosphate uptake in Z. marina (Rubio et al., 2005) and in another seagrass, Posidonia oceanica (Rubio et al., 2018). Consequently, Na+ -coupling seems to be the energization mechanism for the high-affinity uptake of these nutrients in marine angiosperms. Seagrasses are the only vascular plants able to complete their life cycle submerged in marine environments and Z. marina is the most widespread species throughout the temperate northern hemisphere. These plants evolved from land plants towards a marine lifestyle thriving in a high salinity (0.5 M NaCl) and alkaline pH (8.3). Unlike terrestrial plants that take up nutrients through their roots from soils with a low nitrate:ammonium ratio, most seagrasses take up nutrients through their leaves from the bulk seawater with a high nitrate:ammonium ratio. Roots of seagrasses are primarily involved in the uptake of ammonium and phosphate present at significant concentrations in the pores of the sediment in which seagrasses grow (Lee & Dunton, 1999; Terrados & Williams, 1997; Touchette & Burkholder, 2000). Recently an N2-fixing marine bacterium that lives inside P. oceanica root tissue has been described, but the impact of this terrestrial-type nitrogen-fixing symbiosis on seagrasses N nutrition is unknown (Mohr et al., 2021). Non-N2 nitrogen concentration in marine environments is persistently low, in the case of Z. marina's habitat, nitrate ranges from 0 to 8 µM in the seawater column and is almost negligible in sediment porewater (Touchette & Burkholder, 2000). Thus, among other adaptive changes as the salt tolerant H+-ATPase (Muramatsu et al., 2002), in Z. marina the very low plasma membrane Na+ permeability (Fernandez et al., 1999) and the maintenance of the cytosolic Na+ homeostasis due to a Na+/H+ exchanger operating at the plasma membrane (Rubio et al., 2011) allow it to use Na+ as driving ion for high-affinity nitrate uptake.
The recent availability of seagrass genomes reports unique insight into the genomic losses and gains involved in achieving those structural and physiological adaptations required for their marine lifestyle (Lee et al., 2016; Olsen et al., 2016). This knowledge opens the possibility to characterise unique physiological adaptations of seagrass physiology at the molecular level. In preliminary work, we described that the only sequence quoted as high-affinity nitrate transporter (Zosma70g00300.1; NRT2.1) in NCBI and Phytozome databases conserves the transmembrane topology including the major facilitator superfamily (MFS) domains, the “nitrate signature” of all NNP (nitrate–nitrite porter) family members. Additionally, it is phylogenetically more related to the NRT2.5 orthologues from monocots and dicots than to NRT2.1 proteins (Rubio et al., 2019). The sequence similarity of ZosmaNRT2 to other angiosperm NRT2 transporters is below 59%. This suggests that ZosmaNRT2 may have a closer evolutionary relationship to angiosperm NRT2.5 compared with other NRT2 transporters. Therefore, it was hypothesized that ZosmaNRT2 evolved to use Na+ as a driving ion to support high-affinity NO3− uptake in Z. marina (Rubio et al., 2019). Nevertheless, the ability to use Na+ instead of H+ as driving ion in any of the NRT2 transporters has not been functionally characterised to date (Wang et al., 2018).
The first high-affinity nitrate transporters characterised at the molecular level in vascular plants were NRT2.1 and NRT2.2 from barley and Arabidopsis (Filleur & Daniel-Vedele, 1999; Filleur et al., 2001; Trueman et al., 1996). Thereafter, seven NRT2 proteins (AtNRT2.1–AtNRT2.7) were reported in A. thaliana (Kotur et al., 2012), and other NRT2-like genes have also been cloned and characterised from vascular plants, mosses, fungi, algae and bacteria (Charrier et al., 2015; Fan et al., 2017; Fukuda et al., 2015; Higuera et al., 2016; Machín et al., 2004; Pérez et al., 1997; Tsujimoto et al., 2007). Many NRT2 family members require a partner protein NAR2 (nitrate assimilation-related protein), forming a two-component nitrate uptake system to transport NO3− (Kotur et al., 2012; Miller et al., 2007). In the case of seagrasses, the only NRT2 being isolated is ZosmaNRT2, whose localization at the plasma membrane seems to be stabilized by ZosmaNAR2 (Rubio et al., 2019), but its NO3− transport capacity and kinetic properties have not yet been described.
The other protein family involved in vascular plant nitrate uptake is the NPF family. There are 53, 93 and 331 NPF genes in Arabidopsis, rice and wheat, respectively (Buchner & Hawkesford, 2014; Léran et al., 2014; Li et al., 2021; Wang et al., 2018). Surprisingly, the NPF family displays broad substrate specificity, including nitrate, nitrite, chloride, auxin, ABA and dipeptides, among others (Wang et al., 2018). Most of them display low affinity for nitrate, except Arabidopsis CHL1/NRT1.1/NPF6.3 and rice OsNRT1.1B/OsNPF6.5, which display dual affinities in response to fluctuations in external nitrate concentrations. Indeed, these two members switch between high (micromolar) and low (millimolar) affinity in a process modulated by phosphorylation of a key threonine residue (Liu & Tsay, 2003; Wang et al., 2018). Initially, it was found that AtNPF6.3 was involved in mediating nitrate uptake by the roots as well as root-to-shoot nitrate transport in Arabidopsis (Liu et al., 1999; Léran et al., 2014). As in the case of NRT2 transporters, electrophysiological studies in Xenopus oocytes expressing either NPF6.3 or NRT2s showed that those transport systems from land plants operate as proton-coupled nitrate transport mechanisms (Liu et al., 1999, Liu & Tsay, 2003; Tong et al., 2005; Tsay et al., 2007). Interestingly, functional and structural studies have demonstrated that NPF6.3 functions as a sensor to trigger the primary nitrate response. It is a central component in nitrate-signalling pathways in A. thaliana, regulating many physiological and development aspects (Ho et al., 2009; Wen & Kaiser, 2018). In the marine environment, where nitrate concentrations are persistently low and without significant fluctuations (Pasqueron de Frommeurvault et al., 2016), the dual-affinity nature of NPF6.3 transporter may have been modified, but its role as nitrate sensor cannot be ruled out in seagrasses.
The availability of genetic tools in Z. marina offers the possibility to isolate and functionally characterise the putative Na+-dependent high-affinity nitrate transport in heterologous systems of physiological importance in seagrasses. Thus, the high-affinity nitrate transporter-deficient mutant (∆ynt1) from the yeast Hansenula polymorpha appears to be a useful choice as a heterologous expression system for this purpose. This yeast shows rapid growth in simple defined media, is able to assimilate nitrate as sole nitrogen source (Siverio, 2002), possesses a single high-affinity nitrate transporter that is disrupted in the ∆ynt1 mutant (Pérez et al., 1997), and maintains an inwardly directed Na+ electrochemical gradient by the Nha Na+ (K+)/H+ antiporter and Na+ (K+)-efflux ENA ATPase. These properties are crucial for cytosolic sodium homeostasis at high external pH values (Ramos et al., 2011). Functional complementation of the disrupted strain Δynt1 had been successfully achieved for the expression of the Arabidopsis dual-affinity nitrate transporter CHL1/AtNRT1.1/AtNPF6.3 (Martín et al., 2008) and recently for the plasma membrane nitrate transporter AtNPF6.2/NRT1.4 (Morales de los Ríos et al., 2021).
The aim of this work was to investigate the molecular identity of the Na+ -dependent high-affinity NO3− uptake mechanisms operating at the plasma membrane of the seagrass Z. marina. We undertook this by functional complementation of the high-affinity nitrate transporter gene disrupted strain (Δynt1) of the yeast Hansenula polymorpha. As previously reported for ZosmaNRT2 and ZosmaNAR2 (Rubio et al., 2019) we have isolated the cDNA sequence of the putative high-affinity nitrate transporter ZosmaNPF6.3 from leaves of a natural Z. marina population and analysed the effect of N-starvation on the expression levels of all these sequences, since the Na+-dependent high-affinity NO3− uptake was known to be inducible under these conditions (García-Sánchez et al., 2000). Then, we investigated the NO3− transport capacity, kinetic characteristics, and whether these transporters use Na+ as a driving ion by the functional expression of ZosmaNRT2, ZosmaNRT2::ZosmaNAR2 and ZosmaNPF6.3 in the ∆ynt1 H. polymorpha yeast strain.
2 MATERIALS AND METHODS
2.1 Plant material, treatments and RNA extraction
Z. marina L. plants were collected from Cádiz Bay (36°29′25.9” N, 6°15′49.0” W, Spain) and transported within 2 h to the laboratory in pots containing natural seawater (NSW) at 15°C. Surface epiphytes and older leaves were removed, then plants were maintained in plexiglass containers filled with filtered (0.2 µm) and aerated NSW at 15°C, under a photon flux density (400–700 nm) of 150 µmol m−2 s−1 (16/8 h light/dark photoperiod). Plants were used for experiments within 2 weeks after sampling, renewing the seawater every 3 days.
For gene expression assays, plants were incubated in filtered and aerated NSW (pH 8.3) or artificial seawater (ASW) containing (in mM) 500 NaCl, 55 MgSO4, 12 CaCl2, 10 KCl, 3 NaHCO3, 0.01 NaH2PO4, buffer to pH 8.3 (10 mM Bistris Propane-MOPS) with 10 µM NaNO3 (ASW + N) or without NO3− source (ASW-N). Plants were incubated for 3 h, 3 and 6 days with daily renewal of the medium. Only healthy leaves of Z. marina plants were used for molecular assays and quantitative PCR analysis. Leaves were frozen in liquid nitrogen and stored at −80°C until later use. Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen). RNA was quantified using a NanoDrop™ OneC Spectrophotometer (Thermo Fisher Scientific) and RNA integrity qualitatively checked on a 1% agarose gel. Then, 1 µg of total RNA was used for cDNA synthesis using iScriptTM cDNA Synthesis kit (Bio-Rad).
2.2 Sequence and phylogenetic analysis
As we previously described for ZosmaNRT2 (Zosma70g00300.1) sequence analysis (Rubio et al., 2019), BLASTp searches using Phytozome v12.1 search tool was used to find the putative auxiliary NRT2 protein NAR2 and the putative dual-affinity nitrate transporter protein CHL1/NRT1.1/AtNPF6.3 in Z. marina genome using A. thaliana NAR2 (At5g50200) and NPF6.3 (At1g12110.1) as queries, respectively. Multiple sequence alignments (MSA) on the entire sequences including dicots, monocots, seagrasses, lycophytes, bryophytes and algal proteins were performed by MultAlin alignment tool (http://multalin.toulouse.inra.fr) (Corpet, 1988). Neighbour-joining method (Saitou & Nei, 1987) was used to generate a phylogenetic tree using the bootstrap resampling analysis (1000 bootstrap replicates) included in the SeaView4 programme (Gouy et al., 2010). FigTree v1.4.4 software (http://tree.bio.ed.ac.uk/software/figtree/) was used for visualization. The TMHMM 2.0 server (Krogh et al., 2001) was used for predictions of putative transmembrane domains (TMDs). The online PROTTER application was used for graphic representation of protein topology.
2.3 Analysis of gene expression
Gene expression was determined by quantitative real-time PCR using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) on a CFX96 real-time PCR system (Bio-Rad). The PCR reaction included 6 ng of cDNA in a total volume of 10 μL. The reactions were initially denatured (95°C/30 s), then subjected to 40 cycles of 95°C/15 s, 60°C/30 s. The analysis was carried out using three biological replicates per tissue and per treatment, with at least three technical repeats for each cDNA sample. As internal controls, the housekeeping genes EloF1D (Zosma10g01390), GADPH (Zosma211g00170) and TBP (Zosma425g00050.1) were used. Primer sequences for all genes are available in Supporting Information: Table 1. Primer specificity for each gene was evaluated by PCR from cDNA as template and then visualized through agarose gel electrophoresis. Genes were quantified by the method (Livak & Schmittgen, 2001).
2.4 Isolation and cloning of high-affinity nitrate transporters from Z. marina leaves
Coding sequences (CDS) of ZosmaNRT2 (Zosma70g00300.1), ZosmaNAR2 (Zosma63g00220.1) and ZosmaNPF6.3 (Zosma170g00490.1) were amplified from Z. marina leaf cDNA using the primers listed in Supporting Information: Table 1. Then, we obtained pYNR-EX-ZosmaNRT2 and pYNR-EX-ZosmaNPF6.3 by inserting the ZosmaNRT2 or ZosmaNPF6.3 CDS into the SpeI site of the H. polymorpha integrative pYNR-EX(LEU) vector, between the nitrate reductase gene (YNR1) promoter and its terminator sequence (Perdomo et al., 2002). pYNR-EX-ZosmaNAR2 was generated by cloning the ZosmaNAR2 CDS within the PstI-SmaI sites of the integrative pYNR-EX(URA) vector between YNR1 promoter and terminator. The TOP10 Escherichia coli bacterial strain was used for routine plasmid propagation. The sequence accuracy of the resulting construct was verified by DNA sequencing (STAB VIDA, Lda).
2.5 Yeast transformation
To test the nitrate transport capacity of ZosmaNRT2 and ZosmaNP6.3, we used the Hansenula polymorpha high-affinity nitrate transporter mutant strain ∆ynt1, derived from NCYC495 (leu2 ura3), as recipient strain. Two versions of integrative vector pYNR-EX bearing LEU2 or URA3 as gene marker and nitrate reductase gene promoter and terminator (YNR1) to drive the heterologous expression were used (Martín et al., 2008; Perdomo et al., 2002). Plasmids pYNR-EX-ZosmaNPF6.3 and pYNR-EX-ZosmaNRT2 were used to transform the ∆ynt1 leu2 URA3 strain. For co-expression of ZosmaNRT2 and ZosmaNAR2, pYNR-EX-ZosmaNRT2 and pYNR-EX-ZosmaNAR2 were sequentially used to transform the ∆ynt1 leu2 ura3 strain. Plasmids were linearized at BstEII in LEU2 or NcoI in URA3 to lead the genomic integration at the LEU2 or URA3 loci. Yeast transformation was carried out by electroporation, following the protocol described previously (Saraya et al., 2012). Single or double, as in the case of pYNR-EX(LEU2)-ZosmaNRT2 and pYNR-EX(URA3)-ZosmaNAR2, prototroph transformant colonies were selected in a synthetic medium containing 0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulphate (YNB; Difco), 2% glucose plus 5 mM NH4Cl as nitrogen source (YGNH), supplemented with 30 mg/mL of leucine or 20 mg/mL of uracil when necessary. Transformants carrying the construct of interest as those used in further assays were checked by PCR, using cDNA as a template from yeast RNA total extraction (data not shown). ∆ynt1 nitrate transport complementation was checked by growth assays based on 0.5 mM NO3− (Morales de los Ríos et al., 2021). Single yeast colonies were grown for 16 h at 37°C, with shaking, in YGNH medium, then cell cultures were diluted to OD660 ~ 1 in synthetic N-free yeast base medium (YNF, 0.17% w/v) with 2% glucose plus 0.5 mM KNO3, supplemented with 25 mM NaCl or 50 mM sorbitol and buffered at pH 6.3 or 8.3 using 20 mM MES/BTP. Yeast growth for 48 h of continuously shaken cultures was expressed as relative ∆OD660 (Biomate 5; Thermo Scientific).
2.6 Nitrate depletion assays
Nitrate uptake capacity of transformant yeasts was determined as extracellular nitrate depletion rate. Before the assays, cells were grown overnight (16 h) at 37°C with shaking in YGNH medium. Then, active growing cells (OD 660 ~ 1.5) were washed and incubated in YNB medium without amino acids, ammonium sulphate or sodium (YNF; Formedium), 2% glucose plus 2 mM KNO3 (YGNO) for 90 min at 37°C with shaking to induce the expression of nitrate transporter genes. Nitrate depletion assays were performed at 37°C with constant gentle agitation, using 20 mg of washed cells (wet weight) in 1 mL of synthetic N-free yeast base (YNF, 0.17% w/v) supplemented with 2% glucose and 25 mM NaCl or 50 mM sorbitol and buffered at pH 6.3 or 8.3 using 20 mM MES/BTP.
Depletion assays were triggered by adding 50 µM KNO3 to 1 mL aliquots of active growing cell cultures (20 mg mL−1). Incubation of each 1 mL aliquots lasted 1, 4, 8, 12, 16, 20, 30, 45, 60, 120 or 180 min; after that time, each aliquot was centrifuged (11 000 rpm, 30 s) to remove the cells before NO3− measurement. External nitrate was monitored by the highly sensitive method for NO3− determination based on NO3− reduction to NO2− by vanadium(III) trichloride and subsequent spectrophotometric (Biomate 5; Thermo Scientific) determination of NO2− by the formation of an azo-pink dye (García-Robledo et al., 2014). The nitrate compensation point was obtained from the nitrate depletion curves as the external concentration for which the net flux of the nutrient was zero (Edwards & Walker, 1983).
2.7 Uptake kinetics assays
Nitrate uptake kinetics were derived from NO3− depletion rates in assays at different initial NO3− concentrations (from 2.5 to 150 µM NO3−), using YNF media supplemented with 25 mM NaCl and buffered at pH 6.3 or 8.3 (20 mM MES/BTP). To analyse the Na+-dependence of the nitrate uptake kinetics, depletion assays were carried out using 50 µM NO3− as initial concentration in YNF media containing increasing Na+ concentrations (from 1 to 25 mM NaCl) and buffered at pH 6.3 or 8.3 (20 mM MES/BTP). For each treatment, net uptake rates were calculated as the slope of the linear phase of the NO3− concentration depletion time course as a function of cell weight. At least four replicates were conducted for each assay.
2.8 Data presentation and analysis
Data are presented as means ± SEM. Number of repetitions (n) is indicated for each experiment. Data were plotted and analysed using GraphPad Prism version 8.2.1. For uptake kinetics analysis data were fitted to the Michaelis–Menten model or to the Edwards and Walker model when NO3− uptake requires a concentration above a threshold value, called the compensation point (Edwards & Walker, 1983). Thermodynamic relationship of hypothetical NO3− transport employing either H+ or Na+ as the coupling ion was calculated as described by García-Sánchez et al. (2000). Student's t test or Tukey's honestly significant difference were used for statistics comparison. The significance level was set at p < 0.05.
3 RESULTS
3.1 Genome identification and expression analysis of high-affinity nitrate transporters in Z. marina leaves
Keyword search in Z. marina (v2.2, Phytozome ID: 324) renders 12 putative sequences for nitrate transporters. Using AtNRT2.1 and AtNAR2 as query in BLASTp searches two of these sequences were identified as the only NRT2 gene (Zosma70g00300.1) and the one encoding its auxiliary protein NAR2 (Zosma63g00220.1) in the Z. marina genome (Rubio et al., 2019). Phylogenetic analyses showed that the ZosmaNRT2 sequence has a higher homology with NRT2.5 proteins from dicots than with other NRT2 proteins (Rubio et al., 2019). Reconstruction of the phylogenetic tree for ZosmaNAR2 protein renders a different branching pattern, including ZosmaNAR2 protein sequence in a single clade, separated from the cluster of terrestrial vascular plants (Supporting Information: Figure 1). In addition, using CHL1/NRT1.1/AtNPF6.3 as query in a BLASTp search, two sequences were identified in the Z. marina genome: Zosma170g00490.1 (62% identity) and Zosma88g00660 (48% identity). Phylogenetic reconstitution includes Zosma170g00490.1, the putative ZosmaNPF6.3, in the monocot terrestrial vascular plants NPF6.3 clade, but on a separate branch. Zosma88g00660, which showed a higher similarity to AtNPF6.4 (63%) appears in a separate group (Supporting Information: Figure 2).
As in the case of ZosmaNRT2 and ZosmaNAR2 genes (Rubio et al., 2019), in this work we have isolated the putative ZosmaNPF6.3 coding sequence from a Z. marina natural population. Using a cDNA sample from Z. marina leaves incubated in NSW as template, one fragment of 1797 bp was obtained by PCR corresponding to ZosmaNPF6.3 CDS. The sequencing results showed an open reading frame for 598 amino acids, the predicted structure of which contains 12 transmembrane domains (TM) with a long cytosolic loop between TM6 and TM7 (Supporting Information: Figure 3A). It contains a PTR2-1 motif (family proton/oligopeptide symporters signature 1) between Gly90 and Arg100, which is well conserved in eukaryotic and prokaryotic proteins involved mainly in the H+ symport of small peptides (Steiner et al., 1995) and in NPF proteins (Okamoto et al., 2003). According to the crystal structure of AtNPF6.3, ZosmaNPF6.3 also features an EXXER motif containing three conserved residues on TM1 (Glu43, Glu46 and Arg47) and the same conserved residues Lys165 on TM4 and Glu474 on TM10, related to proton coupling (Doki et al., 2013; Parker & Newstead, 2014; Sun et al., 2014). ZosmaNPF6.3 shows the amino acids Tyr350, Thr354 and Phe509 in the same position as the residues His356, Thr360 and Phe511 involved in NO3− binding in AtNPF6.3 (Supporting Information: Figure 3B). Nevertheless, His356 is not conserved among plant NPF6.3 orthologues, which harbour either a tyrosine (as occurs in ZosmaNPF6.3) or a hydrophobic amino acid at the equivalent position (Sun et al., 2014; Jacquot et al., 2017). The nitrate-binding pocket in ZosmaNPF6.3 seems to be formed by the same hydrophobic residues (Leu49, Val53, Leu78 and Phe511) as in AtNPF6.3 (Sun et al., 2014). Finally, the strictly conserved Thr among plant NPF6.3 orthologues, whose phosphorylation switches CHL1 from low to high nitrate affinity in Arabidopsis (Liu et al., 1999), is also preserved in ZosmaNPF6.3.
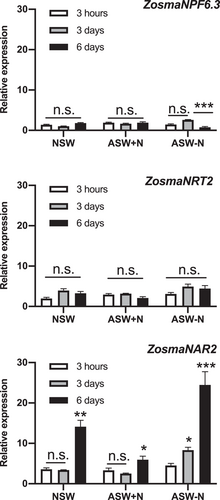
In Z. marina leaf cells, Na+-dependent high-affinity NO3− uptake was inducible by N starvation (García-Sánchez et al., 2000). Thus, to evaluate the expression levels of the potential Na+-dependent NO3− transporters identified in the Z. marina genome, total RNA was isolated from leaves of Z. marina plants incubated for 3 h, 3 and 6 days in natural or artificial seawater containing different NO3− concentrations (NSW: 7.4 ± 0.7 µM, ASW + N: 10 µM and ASW-N: no N added, respectively). qPCR analysis indicated that both ZosmaNRT2 and ZosmaNPF6.3 showed low and similar expression levels despite time and external NO3− concentrations (Figure 1). By contrast, expression levels of ZosmaNAR2 were upregulated in low NO3− treatments, with transcript levels increasing by 2.5-fold in NSW and fourfold in ASW-N (p = 0.0003, p < 0.0001, respectively; two-way analysis of variance [ANOVA]; Tukey Test) after 6 days (Figure 1). Interestingly, the ZosmaNAR2 expression level in natural conditions (NSW), 14.1 ± 1.6 uae; n = 13, was almost 4.4 times higher than the expression of ZosmaNRT2 (3.2 ± 0.5 uae; n = 13). Thus, in Z. marina leaves the results of the relative expression pattern indicate that instead of the specific nitrate transporter genes ZosmaNRT2 and ZosmaNPF6.3, which are expressed at low and time-invariant levels, ZosmaNAR2 is the most expressed gene and would be the main target under low NO3− conditions.
3.2 Characterisation of ZosmaNRT2 and ZosmaNPF6.3 transport activity by heterologous expression in yeast
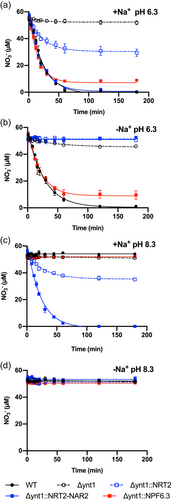
To analyse the NO3− transport capacity of ZosmaNPF6, ZosmaNRT2 and ZosmaNAR2 nitrate-depletion assays were conducted using single transformants ∆ynt1::NPF6.3, ∆ynt1::NRT2, ∆ynt1::NAR2 or the double transformant that co-expresses ZosmaNRT2 and ZosmaNAR2 (∆ynt1::NRT2::NAR2) in media containing 50 µM NO3− as initial concentration. Assays were also performed using H. polymorpha WT and ∆ynt1 mutant strains as positive and negative nitrate uptake controls, respectively. ∆ynt1::NAR2 strain, as well as ∆ynt1 mutant, failed to deplete external NO3− in all assay conditions, indicating that ZosmaNAR2 does not contribute per se to take up NO3− (Supporting Information: Figure 4). WT, ∆ynt1::NPF6.3 and ∆ynt1::NRT2::NAR2 showed similar depletion curves, whereas ∆ynt1::NRT2 showed a lower NO3− depletion capacity in the presence of Na+ at pH 6.3 (Figure 2a). Initial NO3− uptake rates, calculated as the slope of the depletion curves within the first 20 min, were similar for WT, ∆ynt1::NPF6.3 and ∆ynt1::NRT2::NAR2 strains (0.084 ± 0.005; 0.094 ± 0.003 and 0.098 ± 0.02 nmol NO3− min−1 mg cells−1, respectively) and twofold higher than those observed in ∆ynt1::NRT2 (0.039 ± 0.009 nmol NO3− min−1 mg cells−1, n = 5; p < 0.0001, two-way ANOVA, Tukey test). After 60 min of incubation, the depletion curve of ∆ynt1::NRT2 strain was stabilized at a compensation point of 30.4 ± 1.4 µM NO3−, that of ∆ynt1::NPF6.3 reached a lower value (7.1 ± 0.9 µM NO3−) whereas the WT and ∆ynt1::NRT2::NAR2 strains fully depleted external NO3− after 120 min of incubation (Figure 2a, Table 1).
| 25 mM NaCl | 50 mM Sorbitol | ||||
|---|---|---|---|---|---|
| V0 (nmol NO3− mg cell−1 min−1) | PCNO3− (µM NO3−) | V0 (nmol NO3− mg cell−1 min−1) | PCNO3− (µM NO3−) | ||
| ∆ynt1::NPF6 | pH 6.3 | 0.094 ± 0.003a | 7.1 ± 0.9c | 0.075 ± 0.002a | 8.9 ± 1.1c |
| pH 8.3 | n.d. | 52 ± 0.2a | n.d. | 50.4 ± 0.1a | |
| ∆ynt1::NRT2 | pH 6.3 | 0.039 ± 0.009b | 30.4 ± 1.4b | n.d. | 51.4 ± 0.5a |
| pH 8.3 | 0.043 ± 0.001b | 35.4 ± 0.7b | n.d. | 51.3 ± 0.5a | |
| ∆ynt1::NRT2::NAR2 | pH 6.3 | 0.098 ± 0.002a | 0.7 ± 0.6d | n.d. | 51 ± 0.6a |
| pH 8.3 | 0.112 ± 0.005a | 0 ± 0.1d | n.d. | 53.3 ± 0.3a | |
- Note: Initial uptake rates (V0; nmol NO3− mg cell−1 min−1) were calculated as the slope of the linear phase from the depletion curves normalized to the wet weight of the cells in each assay (20 mg). The NO3− compensation points (PCNO3−, µM NO3−) were obtained from the external NO3− concentration at the plateau phase of the depletion curves. Data are mean ± SEM of at least four independent assays. “n.d.” denotes treatments without any depletion of external NO3−. For each parameter, significant differences are indicated by different letters (two-way ANOVA, Tukey's test, p < 0.01).
To study the Na+-dependence of NO3− uptake mediated by ∆ynt1::NPF6.3, ∆ynt1::NRT2 and ∆ynt1::NRT2::NAR2 strains, NO3− depletion assays were conducted in the absence of Na+, replacing 25 mM NaCl by 50 mM sorbitol in the medium to maintain osmolarity. In the absence of Na+, external NO3− was depleted only by H. polymorpha WT or ∆ynt1::NPF6.3 strains (Figure 2b). Both strains showed similar initial uptake rates (0.077 ± 0.016 and 0.075 ± 0.002 nmol NO3− min−1 mg cells−1, in assays using WT and ∆ynt1::NPF6, respectively), but external NO3− was not completely depleted by ∆ynt1::NPF6.3, leveling off at 8.9 ± 1.1 µM NO3− after 120 min. This was a similar compensation point to that observed for this strain at pH 6.3 in the presence of Na+ (Table 1). These results indicate that NO3− uptake mediated by ZosmaNPF6.3 would be a Na+-independent mechanism. On the contrary, since no significant decrease in NO3− concentration was observed in assays using ∆ynt1::NRT2 or ∆ynt1::NRT2::NAR2 strains in the absence of Na+ (Figure 2b), NO3− uptake mediated by ZosmaNRT2 transporter would be expected to be a Na+-dependent mechanism.
To evaluate the role of H+ as driving ion in NO3− uptake by yeasts expressing Z. marina nitrate transporters, assay media were buffered to pH 8.3 in the presence or absence of Na+ (containing 25 mM NaCl or 50 mM Sorbitol, respectively). At alkaline pH in the presence of Na+, only strains expressing ZosmaNRT2 were able to take up NO3−. Transformant strains co-expressing ZosmaNRT2 and ZosmaNAR2, unlike ∆ynt1::NRT2, was the sole strain able to fully deplete external NO3−. The ∆ynt1::NRT2 depletion curve reached a compensation point of 35.4 ± 0.7 µM NO3− after 120 min of incubation (Figure 2c). Furthermore, the ∆ynt1::NRT2::NAR2 strain showed 2.6-fold higher initial NO3− uptake rate (0.112 ± 0.005 nmol NO3− min−1 mg cells−1) than observed in the single ∆ynt1::NRT2 strain (0.043 ± 0.001 nmol NO3− min−1 mg cells−1; p < 0.0001, two-way ANOVA, Tukey test) (Table 1). These results indicate that co-expression of ZosmaNRT2 and ZosmaNAR2 increases NO3− uptake capacity, but no significant differences were found in initial NO3− uptake rates observed in the presence of Na+, despite the external pH, suggesting that ZosmaNRT2 drives a Na+-dependent and H+-independent NO3− uptake mechanism. Finally, none of the strains caused the external NO3− concentration decrease in assays performed at pH 8.3 in the absence of Na+ (Figure 2d). This leads to the conclusion that an ion driving force, either due to H+ or Na+, is required for nitrate transport at the micromolar range (50 µM) by H. polymorpha WT or those expressing Z. marina NO3− transporters.
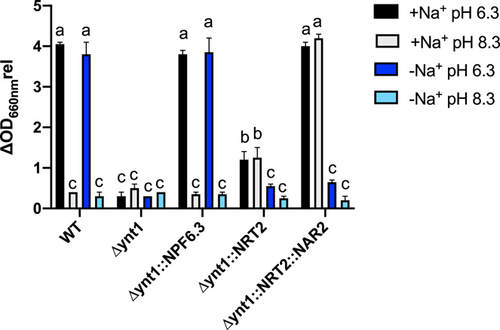
Since no significant growth was observed in either control or transformant yeast strains grown on media containing 50 μM NO3− (data not shown), the nitrate growth complementation capacity of ∆ynt1 transformants expressing ZosmaNPF6.3 (∆ynt1::NPF6.3), ZosmaNRT2 (∆ynt1::NRT2) or co-expressing ZosmaNRT2 and ZosmaNAR2 (∆ynt1::NRT2::NAR2) was evaluated in media containing 500 µM NO3−. The ∆ynt1::NPF6.3 strain showed similar growth behaviour than the WT strain, which only grew at acidic pH regardless of external Na+ (Figure 3). On the contrary, strains expressing ZosmaNRT2 grew in media containing Na+ but showed a partial growth capacity, ≈ 30% of that observed in the WT strain. Interestingly, in Na+ -containing media at both acidic and basic pH, ∆ynt1::NRT2::NAR2 showed similar relative growth to that of WT (Figure 3). These results indicate that to restore the impaired growth capacity, transformant strain expressing ZosmaNPF6.3 required acidic pH while transformants expressing ZosmaNRT2 required the presence of Na+ regardless of the external pH.
3.3 Saturation kinetics of NO3− uptake mediated by ZosmaNRT2 and ZosmaNPF6
To study the kinetics of nitrate uptake mediated by the Z. marina nitrate transporters ZosmaNRT2 and ZosmaNPF6.3, we examined the nitrate uptake rates of ∆ynt1::NRT2; ∆ynt1::NRT2::NAR2 and ∆ynt1::NPF6.3 strains from depletion assays at different initial NO3− concentrations (from 2.5 to 150 µM NO3−). Nitrate uptake rates were calculated as the slope of its depletion curves during the first 20 min of incubation, in media containing 25 mM Na+ and buffered to pH 6.3 or 8.3.
The ∆ynt1::NPF6.3 strain failed to uptake NO3− below 5 µM at pH 6.3 and apparently showed dual NO3− uptake kinetics (Figure 4a). Below 50 µM, NO3− uptakes rates were saturated; fitting the values to the Edwards and Walker model yielded a NO3− compensation point of 5.2 ± 0.3 µM NO3−, half the KM value of 11 ± 2.5 µM NO3− (Table 2). At higher external NO3−, uptake rates increased but were not saturated. In assays at pH 8.3, ∆ynt1::NPF6.3 transformant did not uptake NO3− at any external NO3− concentration (Figure 4a). In contrast, the ZosmaNRT2-expressing transformants were able to take up NO3− in the micromolar NO3− concentration range tested, showing saturation kinetics fitting the Michaelis–Menten equation, regardless of external pH (Figure 4b). In addition, pH had no observed effect on the kinetic parameters of either transformant expressing ZosmaNRT2 (Table 2). However, at both pH 6.3 and pH 8.3, the transformant strain co-expressing ZosmaNRT2 and the auxiliary protein ZosmaNAR2 showed twice the Vmax (0.12 vs. 0.06 nmol NO3− min−1 mg cells−1) and almost six times lower than KM (≈5 vs. 27 µM NO3−), than the single transformant ∆ynt1::NRT2 (Table 2). The ∆ynt1::NRT2::NAR2 strain showed similar KM and Vmax values at pH 6.3 (5.8 ± 0.8 µM NO3−; 0.121 ± 0.004 nmol NO3− min−1 mg cells−1) and at pH 8.3 (5.6 ± 0.5 µM NO3−; 0.121 ± 0.002 nmol NO3− min−1 mg cells−1), respectively. In summary, these results strongly indicate that ZosmaNRT2-mediated high-affinity nitrate uptake is a H+-independent, Na+-dependent transport mechanism and co-expression with ZosmaNAR2 increases the number of active transporters and raises the affinity for NO3−.
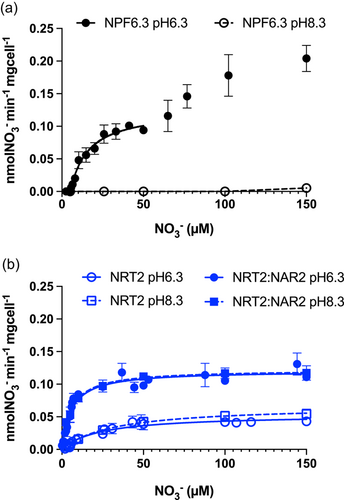
| NO3− Kinetic parameters | pH 6.3 | pH 8.3 | ||
|---|---|---|---|---|
| KM (µM NO3−) | Vmax (nmol NO3− mg cell−1 min−1) | KM (µM NO3−) | Vmax (nmol NO3− mg cell−1 min−1) | |
| ∆ynt1::NPF6.3 | 11.1 ± 2.5b | 0.126 ± 0.009a | n.d. | n.d. |
| ∆ynt1::NRT2 | 24.5 ± 4.7a | 0.053 ± 0.004b | 31.5 ± 2.1a | 0.067 ± 0.001b |
| ∆ynt1::NRT2::NAR2 | 5.8 ± 0.8c | 0.121 ± 0.004a | 5.7 ± 0.5c | 0.122 ± 0.002a |
| Na+ Kinetic parameters | pH 6.3 | pH 8.3 | ||
|---|---|---|---|---|
| KM (mM Na+) | Vmax (nmol NO3− mg cell−1 min−1) | KM (mM Na+) | Vmax (nmol NO3− mg cell−1 min−1) | |
| ∆ynt1::NRT2 | 1.1 ± 0.2a | 0.043 ± 0.001b | 0.87 ± 0.16a | 0.045 ± 0.001b |
| ∆ynt1::NRT2::NAR2 | 0.98 ± 0.8a | 0.099 ± 0.002a | 0.92 ± 0.1a | 0.108 ± 0.003a |
- Note: For NO3− kinetics, data from Figure 4 were fitted to the Edwards and Walker equation for assays using ZosmaNPF6.3 transformants and to the Michaelis–Menten model for kinetics using ZosmaNRT2 or ZosmaNRT2::ZosmaNAR2 transformants and the KM and Vmax. In the case of Na+ kinetic, data from Figure 5 were fitted to the Michaelis–Menten model and the KM and Vmaxvalues calculated. Values are presented as mean ± SEM of at least four independent assays. “n.d.” denotes treatments with no NO3− uptake kinetics. For each ion and parameter, significant differences are indicated by different letters (two-way ANOVA, Tukey's test, p < 0.01).
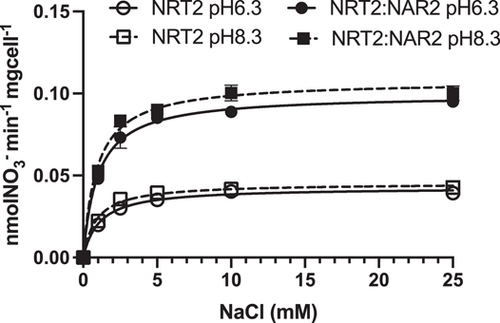
To further characterise the Na+-dependence of high-affinity NO3− uptake mediated by the ZosmaNRT2 transporter, we examined the NO3− uptake rates in response to the saturating NO3− concentration (50 µM NO3−) over a range of mM Na+ concentrations (from 1 to 25 mM). As shown in Figure 5, both the single ZomaNRT2 transformant strain and the double transformant co-expressing ZosmaNRT2 and ZosmaNAR2 showed Na+ saturation kinetics in assays at pH 6.3 or pH 8.3. Regardless of the external pH, NO3− uptake was saturated at 5 mM Na+, fitting the data with the Michaelis–Menten model resulted in similar half-saturation value for Na+ (≈1 mM Na+). On the other hand, the ∆ynt1::NRT2::NAR2 strain showed a twofold higher maximum uptake rate than the single ∆ynt1::NRT2 transformant (Table 2). These results suggest that Na+ coupling would be a specific feature of ZosmaNRT2 transporter, rather than dependence on the presence of ZosmaNAR2, whose function appears to be to increase the number of active ZosmaNRT2 transporters in the membrane and raises its affinity for NO3− (Figure 6).
4 DISCUSSION
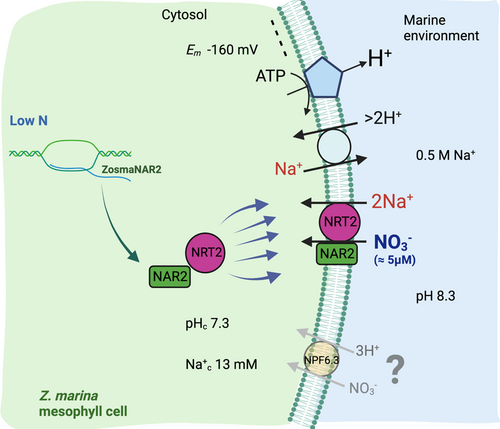
Our group reported the first physiological evidence of a Na+-dependent high-affinity NO3− uptake mechanism operating at the plasma membrane of the marine vascular plant Z. marina (García-Sánchez et al., 2000; Rubio et al., 2005). This has also been proved to function for other nutrient, that is, Pi and amino acid, and in another seagrass, P. oceanica (Rubio et al., 2018), suggesting it is a key mechanism in the adaptation of vascular plants to the marine environment. Seagrasses are the only angiosperms that live completely submerged in the marine environment, facing a persistently low nitrate concentration (≈5 µM NO3−) and showing approximately half the (inward directed) electrochemical potential gradient for H+ than for Na+, due to the inside negative membrane potential (≈−170 mV), the alkaline pH (8.3) and the high Na+ concentration (0.5 M) of seawater (Rubio & Fernández, 2019). However, the molecular identity of the transporter which has acquired a mechanism using the electrochemical Na+ gradient instead of the H+ gradient to drive the high-affinity NO3−uptake in marine vascular plants has remained unknown so far.
To identify the molecular basis of this mechanism, we started by using homology search for high-affinity nitrate transporters among vascular plants, which showed two possible candidates in the Z. marina genome: ZosmaNRT2.5, the only conserved homolog of the high-affinity nitrate transporters belonging to the NRT2 family (Rubio et al., 2019) and ZosmaNPF6.3, the closer homolog to AtNPF6.3, also known as CHL1, the first plant nitrate transporter expressed and characterised by heterologous expression in yeast (Martín et al., 2008). The Z. marina genome also contains only one sequence coding ZosmaNAR2, the partner protein of NRT2 required for the two-component high-affinity NO3− transport system that has been subsequently demonstrated in members of the Archaeplastida from the chlorophyte alga Chlamydomonas reinhardtti (Quesada et al., 1994) to the vascular plants Arabidopsis (Kotur et al., 2012; Okamoto et al., 2006; Orsel et al., 2006), rice (Araki & Hasegawa, 2006; Cai et al., 2008; Yan et al., 2011), barley (Ishikawa et al., 2009; Tong et al., 2005), wheat (Cai et al., 2007) or pepper (Lizama-Gasca et al., 2020).
Olsen et al. (2016) argue that Z. marina's genome indicates that some adaptation mechanisms seem to arise from changes in the same gene families, rather than from speciation of pre-existing genes. However, the latter appears to be the case for the nitrate uptake mechanism in this species. Thus, contrasted with terrestrial plants, where several members of both the NRT2 and NAR2 families are present (Zoghbi-Rodríguez et al., 2021), the two-component model for the high-affinity NO3− uptake seems to be simplified in Z. marina, perhaps a result of its adaptation to the marine environment. ZosmaNRT2 is separated on the phylogenetic lineage from A. thaliana NRT2 proteins, except for AtNRT2.5. This is the only one that shares a common ancestor with monocots, detached from the rest of the NRT2 family transporters (Rubio et al., 2019). Interestingly, ZosmaNAR2 appears in an isolated branch, separates from the reported NAR2 protein divergence between monocot and dicot plants (Cai et al., 2008; Guo et al., 2020; Okamoto et al., 2006), which might suggest a modified function. ZosmaNAR2 apparently plays a role in targeting ZosmaNTR2.5 to the plasma membrane (Rubio et al., 2019) and conserves the Asp104 (equivalent to residue Asp105 in AtNAR2.1), known to be essential for high-affinity NO3− uptake in A. thaliana (Kawachi et al., 2006).
Phylogenetic analysis groups ZosmaNPF6.3 into the monocot clade, sharing a common ancestor with NPF6.3 proteins in dicots. ZosmaNPF6.3 protein topology shows the same number of TMDs as other NPF6.3 orthologues from microalgae, bryophytes, lycophytes and angiosperms, with a long cytosolic loop between TM6 and TM7, indicating it is a well-conserved protein. In addition, the amino acid sequence of ZosmaNPF6.3 contains the characteristic EXXER motif related to proton coupling and key residues at the same position for a nitrate-binding pocket as AtNPF6.3 (Doki et al., 2013; Parker & Newstead, 2014; Sun et al., 2014). ZosmaNPF6.3 also has a Thr103, that could be related to the Thr residue conserved among plant NPF6.3 orthologues, which phosphorylation (Thr101) by CIPK23 kinase switches on the function of NPF6.3 as a high-affinity nitrate transporter in A. thaliana (Ho et al., 2009; Liu & Tsay, 2003).
The qPCR analysis carried out in Z. marina plants incubated in the absence of NO3− showed that neither ZosmaNRT2 nor ZosmaNPF6.3 significantly changed their expression from 3 h to 6 days, compared with control conditions (NSW or ASW containing 10 µM NO3−). These results indicate that both ZosmaNRT2 and ZosmaNPF6.3 show a low but constitutive expression that is not regulated by external NO3−, while ZosmaNAR2 shows the highest expression in NSW and almost double that in the absence of NO3−. This expression pattern differs from that in Arabidopsis, where AtNAR2.1 was induced sixfold by 1 mM KNO3 (Okamoto et al., 2006), whereas the expression of AtNRT2.5 was strongly suppressed by nitrate (Okamoto et al., 2003). The pattern also differs from the members of the Poaceae in the Poales such as rice, where OsNAR2.1 and OsNAR2.2, as well as the four OsNRT2 (OsNRT2.1-4), were upregulated in the roots of N-starved plants (Cai et al., 2008). Indeed, barley displayed co-expression patterns of eight HvNRT2 genes and two HvNAR2 under low nitrate conditions (Guo et al., 2020). Interestingly, ZosmaNAR2 showed a similar low expression to ZosmaNRT2 in ASW containing 10 µM NO3−. Compared with NSW, where NO3− concentration was lower than 5 µM, down-regulated expression of ZosmaNAR2 in response to a slight rise in external NO3− suggests that ZosmaNAR2 functions as the main target of NO3− levels rather than ZosmaNRT2. Expression of the latter diverges from that of its homolog AtNRT2.5, the main high-affinity NO3− transporter induced under nitrogen starvation (Lezhneva et al., 2014). Thus, the inducible high-affinity NO3− uptake reported in 3 days N-starved Z. marina plants (García-Sánchez et al., 2000) could be explained by an increase in ZosmaNAR2, the ZosmaNRT2 partner protein, which promotes an increase in the number of transporters present in the plasma membrane (Rubio et al., 2019). In summary, these results support the two-component system ZosmaNRT2/ZosmaNAR2 being involved in the nitrate deprivation response more than ZosmaNP6.3 in Z. marina plants, providing the high-affinity NO3− uptake in marine environments. However, because ZosmaNPF6.3 could rapidly switch between high- and low-affinity nitrate uptake at the posttranslational level, its contribution cannot yet be ruled out.
To address this issue, we used a high-affinity nitrate transport-deficient strain of H. polymorpha (∆ynt1) for heterologous expression of ZosmaNPF6.3 as well as the two-component system ZosmaNRT2/ZosmaNAR2. The ∆ynt1::NPF6.3 transformant failed to take up NO3− at external NO3− concentrations lower than 5 µM, while at higher concentrations, NO3− uptake was only observed at acidic external pH values. In these conditions, NO3− uptake mediated by ZosmaNPF6.3 showed saturation kinetics with a KM of 11 µM NO3−, which is fivefold higher than that reported in Z. marina leaf mesophyll cells (KM = 2.3 µM; García-Sánchez et al., 2000). In addition, ZosmaNPF6.3 NO3− transport was independent of the presence of Na+ in the medium. The above evidence points to ZosmaNPF6.3 functioning as a proton/nitrate-coupled symporter and rules it out as the Na+-dependent high-affinity NO3− uptake system operating in Z. marina. However, ZosmaNPF6.3 could play other roles in seagrasses, as in Arabidopsis (Noguero & Lacombe, 2016). Similar pH dependence for NO3− transport to that observed here in ZosmaNPF6.3 has been demonstrated in several follow-up studies of NPF6.3 transporters. One of the most well-studied, AtNPF6.3, showed a NO3−- and pH-dependent current when expressed in Xenopus oocytes, indicating that AtNPF6.3 encodes an electrogenic, proton-coupled, NO3−transporter (Liu & Tsay et al., 2003). Analogous results have been reported for the rice homolog, OsNRT1.1a/OsNPF6.3a, which showed lower nitrate transport when external pH rose from pH 5.5 to pH 8.4 (Lin et al., 2000). Furthermore, a nonbiphasic high-affinity nitrate transporter, the maize homolog ZmNPF6.6, also showed pH-dependent NO3− transport in oocyte assays (Wen et al., 2017). In addition to its proton-coupled nitrate transport activity (Tsay, 1993), NPF6.3 functions as a nitrate sensor in terrestrial plants. It is also able to promote physiological responses in the control of root system architecture and modulate expression levels of many genes implicated in nitrate signalling pathways (Krouk et al., 2010, 2010b; Medici & Krouk, 2014; Medici et al., 2015). In terrestrial plants, NPF6.3 dependent regulation of nitrate transporters is related to nitrate concentration (Noguero & Lacombe, 2016). Thus, fast induction of CHL1 (NPF6.3) expression is observed in response to brief exposure to nitrate (Ho et al., 2009), whereas NRT2.1 is down-regulated with long-term high nitrate supply (Krouk et al., 2006; Muños et al., 2004; Laugier et al., 2012). Seagrasses thrive in N-deprived natural systems, typically <5 µM, and nitrate requirements for their growth are generally saturated below 4 µM (Zimmerman et al., 1987). Interestingly, the affinity for NO3− of ZosmaNPF6.3 (11 µM NO3−), which is in the high-affinity NO3− uptake range of terrestrial plants (6–100 µM, Crawford & Glass, 1998), is higher than the specific affinity reported for AtNPF6.3 with a KM ranging between 50 and 80 µM (Liu et al., 1999; Sun et al., 2014). This suggests that ZosmaNPF6.3 is able to transport NO3− at very low concentrations of this ion, although, the H+-dependence points to a minor role of ZosmaNPF6.3 as a contributor for the high-affinity nitrate uptake mechanism in Z. marina. Interestingly, a key feature of NPFs is that H+-coupling appears to be separated from substrate recognition, providing them with flexibility in substrate selectivity (Parker et al., 2017). AtNPF6.3 maize homologs ZmNPF6.6 and ZmNPF6.4 and Medicago truncatula MtNPF6.5 transport NO3− but also mediate Cl− uptake with different preference when expressed in Xenopus (Wen et al., 2017; Xiao et al., 2021). Recently, it has been proposed that in the absence of NO3− AtNPF6.3 could play a central role in Cl− uptake from the environment (Liu et al., 2020) suggesting that further assays are needed to test whether ZosmaNPF6.3 could mediate a similar function in Z. marina leaves.
Contrary to ZosmaNPF6.3, co-expression of ZosmaNRT2.5/ZosmaNAR2 functionally complemented ∆ynt1 yeast and mimics the physiological characteristics, that is, NO3− high-affinity and Na+-dependence of the high-affinity NO3− uptake, observed in Z. marina leaves (García-Sánchez et al., 2000). This suggests that these two components of the NO3− high-affinity transport system evolved in seagrasses, allowing NO3− uptake in the marine environment (Figure 6).
Single expression of ZosmaNRT2 partially restored high-affinity nitrate uptake in the H. polymorpha ∆ynt1 mutant strain, contrary to previously reported results for all AtNRT2s in Xenopus oocytes except AtNRT2.7, which requires co-expression of AtNAR2s to transport nitrate (Chopin et al., 2007). Later, Kotur et al. (2012) showed that co-injection of all AtNRT2 genes along with AtNAR2.1 resulted in statistically significant increases in 15NO3− uptake by Xenopus oocytes, resulting from single AtNRT2 injections. This would also be the case of the co-expression of ZosmaNRT2 and ZosmaNAR2. The requirement of NAR2 for the high-affinity nitrate transport mediated by NRT2 was initially identified using Chlamydomonas reinhardtii mutants (Quesada et al., 1994). Then, by heterologous expression in Xenopus oocytes, Zhou et al. (2000) demonstrated that only when CrNAR2 was co-expressed with CrNRT2.1 was the high-affinity nitrate uptake observed in Xenopus oocytes. Similar results were reported for barley; the oocytes only took up NO3− when HvNRT2.3 was injected with HvNAR2.3 (Tong et al., 2005). Using rice, OsNAR2.1 interacted with several OsNRT2 transporters promoting NO3− uptake in oocytes as well as in rice, over low to high concentration range (Yan et al., 2011).
Álvarez-Aragón and Rodríguez-Navarro (2017) showed that increasing external NO3− concentration in the millimolar range (0.5–20 mM NO3−) promoted the Na+ accumulation in shoots of A. thaliana hkt1 mutant. In this case, Na+ accumulation depends on the passive root Na+ uptake and its subsequent loading into the xylem, whose return of Na+ from the xylem sap to xylem parenchyma cells is practically zero in the hkt1 mutant. Thus, this NO3− -dependent shoot Na+ accumulation in Arabidopsis seems to be predominantly a charge compensation mechanism, not a NO3− symporter. Na+ flux to the shoot depends on the Na+/H+ antiporter SOS1 activity for xylem loading and does not appear to be coupled to any nutrient uptake mechanism operating at the plasma membrane (Álvarez-Aragón & Rodríguez-Navarro, 2017). However, the results of the present study show how ∆ynt1 functional complementation mediated by ZosmaNRT2 was only observed in the presence of sodium, regardless of external pH. This demonstrates that NO3− transport capacity relies on ZosmaNRT2, irrespective of the presence of ZosmaNAR2, which failed to transport NO3−. Furthermore, the kinetic study indicates that Na+ and NO3− are the real substrates of ZosmaNRT2, since no differences were observed in either KM or Vmax for NO3− and Na+ when ZosmaNRT2 was expressed alone, whatever the external pH. Co-expression with ZosmaNAR2 increases the nitrate maximum uptake rate roughly two and a half-fold. This is similar to that described for 15N accumulation capacity when AtNAR2.1 and AtNRT2s are co-injected into Xenopus oocytes and when AtNRT2.5 is co-injected with AtNAR2.1 (Kotur et al., 2012). In addition, co-expression with ZosmaNAR2 also doubled Vmax in the Na+ kinetics, indicating that in the presence of ZosmaNAR2 the number of active ZosmaNRT2 transporters increases. However, the lack of kinetics assays of the co-expression of NAR2 and NRT2 proteins in heterologous systems does not allow us to compare the increased effect on the nitrate affinity of ZosmaNRT2 when co-expressed with ZosmaNAR2.
Kinetic assays of ZosmaNRT2/ZosmaNAR2 showed a constant value (KM = 5.5 µM) for NO3− semisaturation in the same range as reported previously for leaf mesophyll cells of Z. marina (KM = 2.3 ± 0.78 µM NO3−; García-Sánchez et al., 2000) or P. oceanica (KM 8.7 ± 1 µM NO3−, Rubio et al., 2018). Furthermore, regardless of external pH and single expression of ZosmaNRT2 or ZosmaNRT2/ZosmaNAR2 co-expression, the Na+-dependence of NO3− transport also shows a similar KM value for Na+ (KM = 1 mM) to that reported in Z. marina leaf cells (KM = 0.78 ± 0.18 mM Na+), but lower than in P. oceanica (KM = 7.2 ± 1.1 mM Na+, Rubio et al., 2018). This indicates that the transporter works at saturation in the Na+ concentrations in the marine environment (Figure 6).
Functional expression of ZosmaNRT2/ZosmaNAR2 in ∆ynt1 yeast is consistent with previously reported properties of NRT2 transporters: (i) ZosmaNRT2 requires co-expression of the partner protein ZosmaNAR2 to increase high-affinity nitrate transport and (ii) ZosmaNRT2 transport NO3− without the presence of ZosmaNAR2. This observation has been reported for other NRT2 proteins as in all Arabidopsis NRT2 proteins (except AtNRT2.1 that requires AtNAR2 to transport nitrate), which sustain nitrate transport when expressed in Xenopus oocytes (Kiba et al., 2012; Kotur et al., 2012). Another case is the rice transporter OsNRT2.3b, which mediates nitrate influx when expressed alone in Xenopus oocytes although co-injection with OsNAR2.1 did not improve nitrate transport (Feng et al., 2011). However, ZosmaNRT2 functions as a Na+-dependent, high-affinity nitrate transporter, the first H+-independent NRT2 characterised in a vascular plant. Its interaction with ZosmaNAR2 substantially increases both NO3− affinity and NO3− transport rate. In our previous work, we showed that presence of ZosmaNAR2 increases fourfold the amount of ZosmaNRT2 protein in the plasma membrane in Nicotiana benthamiana leaf cells (Rubio et al., 2019). Interaction between the level of NRT2 and NAR2 proteins has already been observed in Atnar2.1 KO mutants, in which the lack of AtNAR2.1 prevents accumulation of AtNRT2.1 protein in the plasma membrane (Wirth et al., 2007; Yong et al., 2010). Before this, Orsel et al. (2006) noted that the plasma membrane localization of AtNRT2.1 increased when AtNAR2 was present and increasing the nitrate transport, but the nitrate affinity was not investigated. Moreover, Laugier et al. (2012) observed that despite high constitutive AtNRT2.1 transcript accumulation in Arabidopsis roots, high-affinity nitrate uptake was down-regulated in the 35 S::NRT2.1 transformants in response to repressive nitrogen treatments, due to the decreased abundance of NRT2.1 and NAR2 proteins. These findings establish the role of NAR2 in regulating the stability and activity of NRT2 transporters, but it is not yet known whether it also causes a change in affinity. One hypothesis could be that given that NAR2 regulates the function of NRT2 by forming a tetramer of ~150 kDa, consisting of two subunits each of NRT2.1 and NAR2.1 (Yong et al., 2010), such an association/dissociation of the NRT2/NAR2 heterooligomer would also involve a change in NRT2's affinity for NO3−.
In summary, the results from this study show the first molecular evidence for a Na+-dependent high-affinity nitrate transporter operating in an angiosperm. They indicate that ZosmaNRT2 evolved in a manner that used Na+ instead of H+ in Z. marina and probably in other seagrasses, such as P. oceanica. Interestingly, matching the physiological evidence of the Na+-dependent high-affinity NO3− uptake was reported for these seagrasses, ZosmaNRT2 requires co-expression of the partner protein ZosmaNAR2, which increases the amount of ZosmaNRT2 at the plasma membrane (Rubio et al., 2019), upregulates its expression in N-starved plants and increases the affinity and the maximum uptake rate of NO3− at the plasma membrane. Therefore, this Na+ -dependent transport mechanism may be an interesting target for further studies to investigate its role in NO3− sensing and modulation of high-affinity NO3− uptake mediated by NRT2. In addition, to explore the capacity of land plants to use Na+-driven NO3− uptake mechanisms could be beneficial, especially in crops grown in K+ deficient, saline and low-nitrate areas.
ACKNOWLEDGEMENTS
The authors thank Guido Jones for English revision and Prof. Fernando Brun for the Z. marina picture used as cover image. This work is included in the framework of Campus de Excelencia Internacional del Mar (CEIMAR). The University of Dundee is a registered Scottish charity, No 051096. This work is dedicated to Professor Miguel Alcaraz in memoriam. This work has been supported by the Research Funds of Malaga University (0837002020 B4-2021-08) and Andalusia Regional Government (GLOCOMA-FEDER-UCA 18-107243) awarded to Lourdes Rubio and José A. Fernández. Malaga University-CBUA finances open access. Lourdes Rubio and José A. Fernández applied for a grant to the Spanish Ministry of Science and Innovation, reference PID2020-118059RB-I00 for this work. The application was dismissed twice. Jordi Díaz-García is beneficiary of doctoral fellowship from the Spanish Ministry of Universities (FPU18/03300). Lourdes Rubio, José A. Fernández and Jordi Díaz-García are members of the RNM176 research group.
Open Research
DATA AVAILABILITY STATEMENT
All lines and data are available upon request.