Metabarcoding reveals marked seasonality and a distinctive winter assemblage of dinoflagellates at a coastal LTER site in the Gulf of Naples
Abstract
Dinoflagellates constitute an abundant and diversified component of marine plankton, mostly associated with stratified conditions typical of late spring through autumn in temperate regions. Yet, difficulties with the identification of many species limit the knowledge of their composition and seasonal succession. Here we use a 3-year V4-18S rDNA metabarcoding dataset (4,366,007 reads, 4650 Amplicon Sequence Variants, ASVs) collected on 48 dates at the Long-Term Ecological Research site MareChiara (LTER–MC) to explore the diversity and temporal distribution of dinoflagellates in the Gulf of Naples (Mediterranean Sea). A large fraction of dinoflagellate ASVs (55%, 68% of the reads) were assigned to 187 species in 85 genera, while 33% (23% of the reads) were attributed to undetermined Syndiniales, and the remainder to unidentified groups. A total of 147 ASVs were assigned at 100% similarity to 135 reference sequences, corresponding to 116 species, 46 of which were putative new records for the Gulf of Naples. Cluster analysis conducted on a normalized dataset of 1199 ASVs identified four sample clusters that corresponded largely to the spring, summer, autumn and winter seasons, respectively, each including samples from all 3 years. Syndiniales were particularly abundant (43.1%) in the winter cluster, whereas Gyrodinium reached higher percentages in the summer and autumn ones (38.6% and 57.8%), Gymnodiniales sensu stricto were more abundant in spring (11.5%) and Gonyaulacales in summer (13.5%). Almost half of the ASVs analysed (561) were significantly associated with one of the four seasonal clusters (p < .05). The winter group of ASVs was particularly rich (348 ASVs) and mainly consisted of naked and parasitic taxa, which are impossible to identify in routine phytoplankton observations in light microscopy. Despite biases mainly ascribed to both the incompleteness of the reference dataset and the limited resolution of the marker gene, the metabarcoding approach has provided new insights into the ecology and distribution of this important component of the plankton, highlighting its marked seasonality along with the existence of a diversified and previously overlooked dinoflagellate community in winter.
1 INTRODUCTION
Dinoflagellates are a morphologically and functionally diverse group of unicellular eukaryotes that are abundant in marine and freshwater ecosystems. They exhibit a broad spectrum of sizes and shapes; many species perform photosynthesis, others are heterotrophic, while mixotrophy seems widespread in this group of protists (Jeong et al., 2010; Stoecker et al., 2017). The majority of dinoflagellates possess two heteromorphic flagella with which they can swim, but there are also several parasitic and endosymbiontic taxa, which lack flagella at least during part of their life cycle (Horiguchi, 2015; LaJeunesse et al., 2018; Suggett et al., 2017).
Large-sized dinoflagellates have attracted scientific interest ever since the first oceanographic expeditions (Schütt, 1895). Especially during the first half of the 20th century, several taxonomic monographs have described numerous taxa in light microscopy (e.g., Jorgensen, 1911; Kofoid & Swezy, 1921; Lebour, 1925). More recent taxonomic investigations have combined ultrastructural studies in electron microscopy of cultured taxa with nucleotide sequencing approaches, thus uncovering considerable additional diversity among dinoflagellates. This is especially the case for many unarmoured taxa (e.g., Daugbjerg et al., 2000; Hoppenrath et al., 2009; Nezan et al., 2014; Reñé et al., 2015). However, detailed molecular analyses are still lacking even for common armoured genera (e.g., Gonyaulax, Dinophysis, Protoperidinium, Tripos) which could hence harbour great hidden diversity. Research on dinoflagellate diversity has gained considerable interest because several species produce toxins that are harmful to humans and marine organisms (Hallegraeff et al., 2021).
Over the last decades, metabarcoding approaches have been applied to uncover the diversity of planktonic communities and their spatial and temporal composition. These investigations, based on specific barcode regions of the 18S ribosomal RNA gene, enable the detection of taxa independent of microscopy methods along with the identification of taxa lacking prominent morphological features. In metabarcoding studies dinoflagellates often dominate marine planktonic communities (e.g., Gran-Stadniczeñko et al., 2019; Le Bescot et al., 2016; Piredda et al., 2017), a result that could be influenced by the high copy number of the rRNA genes, leading to overestimation of their abundance (Liu et al., 2021). In addition, biases due to the low resolution at species level of the selected barcode region and the incomplete reference dataset could limit the representation of the actual dinoflagellate diversity (e.g., Mordret et al., 2018).
In a metabarcoding study carried out on eight sampling dates at the coastal Long Term Ecological Research station (LTER–MC) in the Gulf of Naples (Mediterranean Sea) dinoflagellates constituted a significant fraction of the protist assemblage, with Dinophyceae accounting on average for 41% and Syndiniales for 15% of the total environmental sequences (Piredda et al., 2017). Based on species identification and enumeration with standard light microscopy (LM) dinoflagellates were the second group after diatoms in terms of biomass, with higher values from late spring till summer (Ribera d'Alcalà et al., 2004; Zingone et al., 2019). This prominent component of the plankton communities in the area has remained largely uncharacterized, because it is generally dominated by relatively small taxa, both armoured and unarmoured, impossible to identify at the species- or even genus-level in LM.
Several efforts in the last decades have aimed to fill the knowledge gap on dinoflagellate diversity in the Gulf of Naples through cultivation and in-depth morphological and molecular analyses of, e.g., unarmoured chloroplast-bearing species (Siano et al., 2009), Scrippsiella spp. (Montresor et al., 2003), and other selected species (e.g., Percopo et al., 2013; Puigserver & Zingone, 2002). Here we have used an HTS-metabarcoding approach based on an ASV analysis of the V4 region of the 18S rRNA gene on a 3 years' dataset to gain new insights into the diversity of dinoflagellates by investigating their temporal patterns and identifying the communities that characterize the different seasons of the year.
2 MATERIALS AND METHODS
2.1 Sampling and sample processing
The LTER–MC (40°48.5′ N and 14°15′ E) is located in the Gulf of Naples (Tyrrhenian Sea, Mediterranean Sea; Figure S1). The site has been sampled for physical and chemical parameters and plankton weekly since 1995 (Ribera d'Alcalà et al., 2004; Zingone et al., 2019). Metabarcoding data were gathered on 48 sampling dates over a 3-year time window (2011–2013) alongside CTD measurements for temperature and salinity, concentrations of chl a and inorganic nutrients – nitrate, nitrite, ammonia, phosphate and silicate. For methods related to the physical and chemical parameters see Sabia et al. (2019). Water samples for phytoplankton counts for the same 48 dates were collected with Niskin bottles at 0.5 m, fixed with neutralized formaldehyde at a final concentration of 0.8%, and examined at a Zeiss Axiovert 200 (Zeiss) microscope (magnification 400×) following the Utermöhl method (Edler & Elbrächter, 2010). Phytoplankton biomass was calculated from mean cell biovolumes using the formula proposed by Menden-Deuer and Lessard (2000) for protist plankton.
For DNA extraction, 3 L of surface seawater (sampled with a Niskin bottle at 0.5 m) were filtered on each of two cellulose ester filters (47 mm diameter, 1.2 μm pore-size, EMD Millipore). DNA was extracted using the DNeasy Plant Kit (Qiagen GmbH) following the manufacturer's instructions. The V4 region of the 18S rDNA was amplified and sequenced with Illumina MiSeq; information on the primers and the sequencing protocol are reported in Piredda et al. (2017).
2.2 Denoising and taxonomic assignment
After read quality assessment with FastQC (Andrews, 2010), primers were trimmed from demultiplexed reads using cutadapt (Martin, 2011). Contig assembly and denoising were performed using the relative abundance of each ASV over its total abundance across all samples.
R library (Callahan et al., 2016), with the default parameters described in the MiSeq tutorial, except for the number of accepted mismatches in the overlap region (8; default no mismatches). The denoising procedure resulted in a total of 38,180 Amplicon Sequence Variants (ASVs) for the V4 regions.
Amplicon Sequence Variants were then taxonomically classified with PR2 v4.14 database (Vaulot et al., 2022), supplemented with additional sequences, mostly from taxa regularly found in the GoN; BLAST was set to return a single match at 90% similarity. The highest scoring matches with the best taxonomic resolution were then selected among the returned results. ASV assigned to dinoflagellate reference sequences with similarity ≥97%, query coverage ≥70% of the sequence length and with read abundance >3 were selected and used for the ASV diversity analyses.
The suprageneric classification of ASV in Superclades refers to the phylogenetically organized taxonomic framework presented in Mordret et al. (2018).
2.3 Statistical analyses
For β-diversity analyses, the dinoflagellate ASV matrix was further reduced by removing ASVs that were only present in 1–3 samples and normalized to 12,947 reads with the rrarefy function. One sample (20 December 2011) only consisting of 4 ASVs (290 reads) was excluded. Statistical analyses were performed using vegan R-package (Oksanen et al., 2017). A hierarchical cluster analysis (HCA) on Hellinger-transformed data based on a Bray–Curtis dissimilarity matrix was carried out. Adonis was used to test the statistical robustness of the clusters.
PermANOVA (adonis function in vegan) was used to detect significant associations with the environmental parameters (temperature, salinity, day length, chl a, total inorganic nitrogen, phosphate and silicate). Canonical Correspondence Analysis (CCA) was used to extract and summarize the variation in the communities among samples using the environmental variables statistically selected.
To identify taxa that characterized the different groups of samples resulting from the cluster analysis, the Indicator Value (IndVal) index (func = ‘IndVal.g’) from indicspecies R-package, was calculated with permutational test and using function multipatt(). The function computes the original index IndVal (Dufrene & Legendre, 1997) with modifications proposed by De Cáceres et al. (2010). The IndVal varies between 0 and 1 and is based on two components: ‘A’ is the estimate of the specificity of an ASV based on its exclusive presence in a group of samples, ‘B’ is the estimate of the fidelity of an ASV to all samples of that group. An ASV with IndVal 1 is present only in samples of a group and in all samples of that group.
3 RESULTS
3.1 Dinoflagellate temporal patterns in light microscopy (LM)
Dinoflagellate abundances estimated from LM cell counts over the 3 years of investigation varied between 6.59 × 103 and 1.30 × 106 cells L−1, representing 2.01 ± 1.34% of the total phytoplankton populations (Figure S2A). Abundance values were generally lowest in winter and highest in spring-early summer. The majority of dinoflagellate cells (85.83 ± 15.02% of total dinoflagellate abundance) could not be identified at the species or even at the genus level and were clustered in unarmoured and armoured taxa of different size categories. Among the undetermined groups, small sized (<15 μm) unarmoured cells dominated the dinoflagellate assemblages (65 ± 17.17% of total dinoflagellate abundance). In terms of biomass, dinoflagellates represented a higher fraction of the total phytoplankton (21.21 ± 16.16%), with values ranging between 0.95 and 313.47 μg C L−1 (Figure S2B).
3.2 ASV diversity
The total dinoflagellate V4 dataset contained 5,125,683 reads, corresponding to 16,980 ASVs and accounting for 47% of the total protist reads (55.6% of total ASVs) recorded on the 48 sampling dates at the LTER–MC site. Upon discarding ASVs with less than 97% similarity to known dinoflagellate references and ASVs represented by less than 4 reads, the data set contained 4650 ASVs consisting of 4,366,007 reads (Table S1). Of these, 2552 ASVs (55% ASV; 2,976,440 reads, 68%) were assigned to 187 dinoflagellate species in 85 genera. A large part of the remainder was assigned to other Syndiniales (1541 ASVs, 33%; 1,000,183 reads, 23%), mainly consisting of taxa matching several clades of groups I and II (sensu Guillou et al., 2008). Finally, a relatively low number of ASVs (557 ASVs, 12%; 389,384 reads, 9%) matched reference sequences falling into unidentified suprageneric groups.
Among the ASVs identified to the species level, the most abundant (54% of the reads) were those assigned to unarmoured species of the genus Gyrodinium (mainly G. fusiforme, G. helveticum, G. rubrum and G. heterogrammum), followed by Lepidodinium and armoured species of Alexandrium and Stoeckeria, each with less than 4% of the reads.
The numbers of ASVs assigned to one and the same reference sequence varied considerably, reaching the highest values in the case of the most abundant taxa. In some cases, different ASVs were assigned to the same reference sequence with different similarity values, while in other cases they were assigned to different reference sequences available for the same species, with variable similarity. For example, Alexandrium margalefii was present with 49 ASVs matching a unique reference sequence and Amoebophrya ceratii with 90 ASVs also matching a unique reference sequence, in each case with variable similarity. A total of 91 ASVs matched four distinct reference sequences of Gyrodinium dorsalisulcum. Gyrodinium fusiforme, represented an extreme case, with 480 ASVs matching 29 distinct reference sequences, several of which with 100% similarity.
A total of 147 ASVs were assigned at 100% similarity to 135 reference sequences, corresponding to 116 species (Tables S1 and S2). In most cases, an ASV was assigned unambiguously to one or more reference sequences of a single species. Yet, there were 18 cases in which multiple congeneric species shared identical V4 references (e.g., toxic Azadinium spinosum/non-toxic A. trinitatum, Karenia brevis/K. mikimotoi/K. papilionacea, several species within the genera Dinophysis and Tripos). The same V4 region was at times also shared among species belonging to different, closely related genera, as in the case of the sequence #AF272045 shared by Karlodinium veneficum, Takayama pulchella and Takayama acrotrocha (Table S2).
A total of 64 ASVs were assigned at 100% similarity to 57 reference sequences of 46 species that had never been recorded previously in the area over more than 30 years of detailed investigations of natural samples and culture material in LM and electron microscopy (D. Sarno, M. Montresor and A. Zingone, unpublished data). Three of these new records comprised ASVs assignable each to a reference sequence representing multiple distinct species, namely, Azadinium trinitatum/A. spinosum, Blastodinium mangini/B. navicula and Apocalathium aciculiferum/A. malmogiense, none of which previously recorded in the Gulf of Naples. The new records included 11 parasite (14 reference sequences), namely, Blastodinium spp., Chytriodinium spp., Dissodinium pseudolunula, Ellobiopsis chattonii, Ichthyodinium chabelardi, Euduboscquella spp. and Syndinium turbo, and two symbiont species, i.e., Brandtodinium nutricula (valid name Zooxanthella nutricula) and Pelagodinium bei.
A total of 22 ASV were assignable to reference sequences of 20 potentially harmful species (e.g., of the genera Alexandrium, Azadinium and Dinophysis; Lundholm et al. (2009), see annotations in Table S2), four of which were among the taxa not recorded before in the Gulf of Naples. Several unambiguously identified ASVs corresponded to small-sized species (<15 μm), such as those in the genera Azadinium, Biecheleria (B. sp. NaD22), Biecheleriopsis (B. adriatica) and Heterocapsa (H. pygmaea), the latter with high abundance.
3.3 Temporal patterns
For β-diversity analyses, the dataset was normalized to 12,947 reads per sample, and the sample from 20 December 2011 was removed because of its very low number of reads. Thus, the dataset used here consisted of 1199 ASV with 608,071 reads in 47 samples (Table S3). In the HCA, the samples grouped in four main clusters, each including samples from the 3 years and named by the season of the prevalent samples in it (Figure 1a): the winter cluster included nine samples (116,523 reads) collected from December to February of the 3 years. Next to it appeared the autumn cluster with seven samples (103,576 reads) collected in October and November plus one of July 2013. The spring cluster, the largest one, grouped 15 samples (219,661 reads) collected from late March through June and one of February 2011. Finally, the summer cluster (168,311 reads) grouped 12 samples gathered from July through September. One sample, collected on 7 March 2012, separated from all the others at high-dissimilarity values (>0.75). Adonis test results (R = .11238, p = .05) confirmed that the dinoflagellate species assemblages differed significantly among the seasonal clusters.

Some differences among the seasonal clusters were visible at the level of the major dinoflagellate groups (Figure 1b), defined as ‘superclades’ in the taxonomic framework of Mordret et al. (2018). Samples of the winter cluster were dominated by the parasitic order Syndiniales (43.1% of the reads) which were, at times, also well-represented (18.2%–22.8%) in other seasons and abundant in selected samples (e.g., 6 September, 25 October and 15 November 2011). Within Syndiniales, Group I (mainly clades 1 and 5 sensu Guillou et al., 2008) reached values up to ~90% in the spring and summer clusters and ~ 60% in the winter and autumn clusters, while Group II (mainly clade 6, 10–11 and 26) accounted for about 25% in the latter seasons (Figure S3). Syndiniales were followed in read abundance by unarmoured taxa belonging to the superclade ‘genus Gyrodinium’ (27.8%), which reached highest percentages in the summer (38.6%) and autumn (57.8%) clusters. The spring and summer clusters were the most diversified in terms of superclades, with higher contributions of Gymnodiniales sensu stricto in the spring cluster (11.5%), Gonyaulacales in the summer one (13.5%) and Suessiales (4.4%–5.5%) in both. Prorocentrales were less variable across seasons (2.1%–3%), whereas some groups were only rarely relatively abundant in a few samples (e.g., Dinophysiales on 10 July 2012 and Thoracosphaerales on 18 March 2013).
Among the environmental parameters tested, the PermANOVA analysis selected a subset of four that were significantly associated with dinoflagellate samples: day length, temperature, salinity and total inorganic nitrogen. The first two axes of the CCA, explained 11.7% and 6.7% of the variance, respectively (Figure 2). Samples were arranged following a clear seasonal pattern, with spring–summer assemblages associated with high temperature and day length values, and autumn–winter ones with higher salinity and nutrient levels.
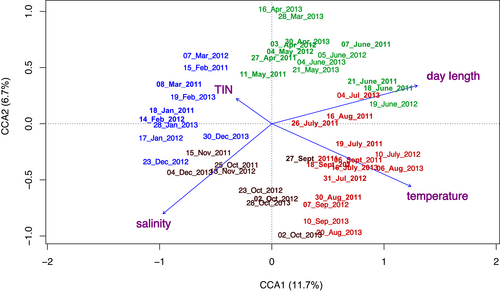
3.4 Species associated with the different seasons
The IndVal analysis identified 561 ASVs as significantly (p < .05) associated with one of the four seasonal clusters, with indicator values ranging between 0.46 and 1.00. A total of 348 ASVs were associated with the winter cluster (238 reference sequences, 72 taxa), 62 with the autumn cluster (45 reference sequences, 32 taxa), 93 with the spring cluster (62 reference sequences, 32 taxa) and 58 with the summer cluster (47 reference sequences, 41 taxa) (Table S4).
A range of different cases existed for the seasonal patterns of individual species represented by multiple ASVs (Table S4). In Pentapharsodinium tyrrhenicum, the five different ASVs matching the reference sequence of the species were all characteristic of the winter cluster (Figure S4A), and the five ASVs matching different reference sequences of Gymnodinium rubrum were all characteristic of the autumn cluster (Figure S4B). Some species, each represented by a large number of ASVs matching the respective reference sequences at ≥97% similarity, e.g., Gymnodinium aureolum (Figure S4C), Prorocentrum cordatum (Figure S4D), were associated with two or more seasonal clusters. An extreme case was Gyrodinium fusiforme (Figure S4E), with 44 ASVs matching 9 distinct reference sequences and associated to all four seasonal clusters irrespective of the reference sequence they matched.
The distribution of the taxa identified by the IndVal analysis across the samples clearly matched the different seasonal clusters (Figure 3, Figure S5). Overall, the groups of species characteristic of the single seasons were rich and diversified. Of the 348 ASVs significantly associated with the winter cluster, 218 belonged to 34 clades of undetermined Syndiniales and matched 174 different reference sequences (Table S4, Figure S5). The most diversified were those belonging to Group I Clade 5 (69 ASVs), Group II Clade 10 and 11 (24) and Group II Clade 6 (24). Among the named ASVs associated with the winter, there were several unarmoured taxa such as Abedinium dasypus, Amphidinium longum, a number of abundant (>100 reads) ASVs of Gymnodinium aureolum (ASV1019 and ASV547), Gyrodinium fusiforme (seven ASVs), G. helveticum (ASV89), Lepidodinium viride (ASV 1364), Paragymnodinium shiwahense (ASV189) and Ptychodiscus noctiluca, as well as armoured taxa such as Pentapharsodinium tyrrhenicum (all five ASVs). Seven of the nine Tripos species with significant IndVal, including T. contrarius and T. limulus/paradoxides, were also associated with the winter samples.
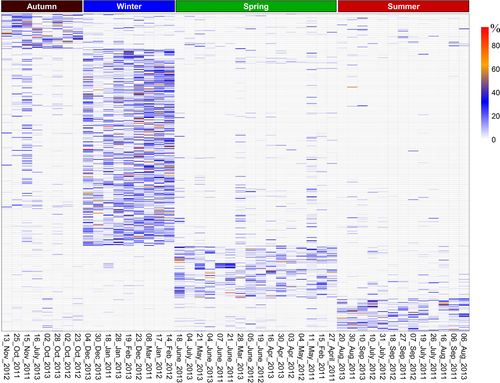
Azadinium dexteroporum and A. trinitatum/spinosum, Alexandrium margalefii, Biecheleria cincta (ASV71 and ASV600), Biecheleriopsis adriatica NaD26, Dissodinium pseudolunula, Gymnodinium aureolum (ASV457) and G. dorsalisulcum (ASV261, ASV359) were among the taxa significantly associated with the spring cluster, while Alexandrium minutum, Fragilidium mexicanum, Protoceratium reticulatum, Pelagodinium bei, Karlodinium/Takayama (ASV407) were among the species associated with the summer one. A small number of taxa were significantly associated with the autumn season, among which Gyrodinium rubrum (ASV24) and Gyrodinium helveticum (ASV453). A variable number of ASVs (16–23) of undetermined Syndiniales were also associated with each of the latter three seasonal clusters.
4 DISCUSSION
4.1 Dinoflagellate diversity
The metabarcoding approach enabled us to uncover a remarkable and so far unknown part of the diversity of dinoflagellates the Gulf of Naples. This diversity includes many abundant taxa that cannot be identified, not even at the genus level, in routine light microscopy observations, as well as rare taxa uncovered thanks to the high-detection power of the method. The most interesting example is represented by Syndiniales, a group of species that spend part of their life cycle as parasites of other dinoflagellates, protists or marine invertebrates, while their free stages are not identifiable with optical methods. The great diversity of this group has been acknowledged only in recent decades thanks to molecular approaches, which have also revealed their abundance in the natural environment in both open oceanic and coastal areas (e.g., Anderson & Harvey, 2020; Armeli Minicante et al., 2019; Clarke et al., 2019; Guillou et al., 2008; Gutierrez-Rodriguez et al., 2022; Sehein et al., 2022; Sogawa et al., 2022). The high-relative abundance of Syndiniales in the winter months in the Gulf of Naples was already noticed in a previous study, where this group of Alveolata accounted for about 50% of the protist community in December (Piredda et al., 2017). In the present study, the marked differences in Syndiniales composition among the seasons are in agreement with the variability in clade dominance reported among different environments and seasons (e.g., Anderson & Harvey 2022; Clarke et al., 2019; Gutierrez-Rodriguez et al., 2022; Sehein et al., 2022). As Syndiniales clades could include many different species, and their abundance presumably reflects the presence of specific hosts, appropriate network analyses involving the whole communities could help unveiling such complex relationships among species inhabiting the water column.
A second very abundant group in our dataset is constituted by a myriad of ASVs attributed to several Gyrodinium species. The dominance and diversity of Gyrodinium spp. are also reported in other metabarcoding studies based on the V4 18S rDNA region. For example, in temperate and tropical locations along the Australian coasts, Gyrodinium accounted for 32% and 82% of the total sequence reads, respectively (Manning et al., 2021), while ASVs attributed to Gyrodinium spirale reached relative abundances spanning between 29.3% and 62.7% in samples collected in spring along the Belgian coasts (Martin et al., 2022). The marked diversity and the dominance of this genus could be the effect of the high number of reference sequences ascribed to this taxon but also reflect the artificial overrepresentation and diversity inflation typical of dinoflagellates that was already noticed in a previous study on eight samples of the dataset analysed here (Piredda et al., 2017), as discussed in the following paragraphs.
While most Syndiniales correspond to the reference sequences of unnamed taxa, mostly known in environmental samples, the majority of the reads in our dataset match the reference sequences of named taxa, in many cases hinting at the possible presence of a large number of species (46 or more if we also included assignments at 99.0%–99.9% similarity) never recorded before in the Gulf of Naples, often not even at the genus level (D. Sarno, M. Montresor and A. Zingone, unpublished data). Even if in the large majority of the cases, there are no other species matching the same reference, it is well known that the V4 fragment used in our study – and at times the whole 18S rDNA region – cannot always discriminate between closely related species (Mordret et al., 2018; Piganeau et al., 2011), and there could still be other unknown species sharing those fragment sequences. Therefore, those newly recorded species should be considered with caution. Nevertheless, these records are a first valuable indication that can stimulate further studies aimed at confirming the presence, in the Gulf of Naples, of those species or of closely related ones by coupling morphological and molecular methods. Not surprisingly, new records include parasite and symbiont taxa (e.g., Blastodinium spp., Chytriodinium spp., Ichthyodinium chabelardi, Syndinium turbo and Pelagodinium bei), which spend most of their life cycle inside other protists or metazoans, or small-sized species of the genera Biecheleria, Biecheleriopsis, Azadinium, Heterocapsa and Karlodinium that can be identified by their thecal plate patterns only using scanning electron microscopy. Among those not seen before are also some species recently described from distant areas in the Pacific, such Yihiella yeosuensis (Jang et al., 2017). As for other ASVs showing 97%–99.74% similarity to reference sequences of previously unrecorded species, they may indicate either local ribotype variants of the same species or different species for which reference sequences are lacking.
The metabarcoding protocol also allows the detection of large species that usually attain low abundances and are underestimated by the Utermöhl method, which is based on the direct examination of small sample volumes. An example is Ptychodiscus noctiluca whose presence in the area has been confirmed upon examining concentrated net samples collected on the dates when the species was abundant in the metabarcoding dataset. The high-detection power of metabarcoding is also evident from the records of the benthic species Ostreopsis cf. ovata as associated with the summer cluster. This species is indeed abundant in that season as epiphyte along the rocky shores of the Gulf of Naples (Migliaccio et al., 2016), but it is rarely observed in net samples at the LTER–MC sampling site which is located 2 nautical miles offshore.
Our study highlights the presence of numerous potentially toxic dinoflagellates, whose distinctive morphological features require either the use of fluorescent stains that allow the visualization of thecal plates and/or the examination in scanning electron microscopy. Azadinium dexteroporum, a species that produces azaspiracid toxins, was first described in the Gulf of Naples based on detailed morphological and molecular investigations of a cultured strain (Percopo et al., 2013); because of its inconspicuous morphological features, only a tentative identification (cf. A. dexteroporum) in LM was possible in some samples. Metabarcoding data confirm its recurrence in the area across the 3 years investigated, with higher read numbers in spring–summer. They also hint at the presence of five other species of the genus, including A. poporum along with Amphidoma languida, which is phylogenetically closely related to the genus Azadinium, both known to produce azaspiracid toxins (Krock et al., 2019; Tillmann et al., 2017).
As already shown in a previous metabarcoding study in the Gulf of Naples (Piredda et al., 2017), Dinophyceae account for a much larger part (47%) of the V4 18S RNA reads compared to their contributions in terms of cell numbers and biomass based on LM data (2% and 21% of the phytoplankton fraction, respectively). This overrepresentation is a well-known effect of the large genome size coupled with the remarkably high number of copies of the nuclear-encoded ribosomal RNA genes in dinoflagellates (Liu et al., 2021; Medinger et al., 2010), which also lead to an artificial inflation of the actual diversity in natural samples. Indeed, even if ASVs are considered to represent the actual diversity more closely than OTUs, a large number of ASVs in our total initial dataset included only a few reads, while many different ASVs were attributed to a single species especially in the case of the most abundant taxa. In some cases, the co-occurrence of these conspecific ASVs over the seasons suggests they are actually variants within the same species. It has recently been shown that the copy number of ribosomal RNA genes significantly correlates with cell size in dinoflagellates (Liu et al., 2021). However, in our dataset large numbers of ASVs are also recorded for medium-sized taxa such as Alexandrium margalefii, suggesting that these multiple ASVs could be the effect of the considerable diversity existing in the structure of dinoflagellate genomes (e.g., Lin et al., 2020; Miranda et al., 2012), which not even the ASV approach is able to dampen.
Contrasting the apparent artificial inflation of dinoflagellate diversity, other cases of ASVs attributed to the same species (with different similarity or based on different reference sequences) but occurring in different seasons of the year suggest that these ASVs may actually belong to different species. As mentioned above, the V4 region of the 18S rRNA gene, although widely applied as metabarcode marker, may not have the appropriate resolution power to tell closely related species apart. In fact, many morphologically distinct dinoflagellate species share similar or even identical marker sequences (Mordret et al., 2018), a limitation that concerns also many other protists (Piganeau et al., 2011). As a matter of fact, the majority of the 320 dinoflagellate species identified in the Gulf of Naples over four decades of observations and by several in-depth studies (D. Sarno, M. Montresor and A. Zingone, unpublished data) have not been detected with the metabarcoding approach adopted in this study. The lack of recovery of many taxa could depend on the scarce resolution of the V4 fragment, besides the limited number of samples examined in this study compared with the whole time series of microscopy data. However, a main hindrance to the recovery of all taxa seems to lie in the incompleteness of the reference sequence databases; V4 references are not yet available for numerous species described over the last centuries based on morphology or, more recently, using molecular markers more variable than the V4 fragment. Less than 20% of the about 2400 described dinoflagellate species are represented by an 18S rDNA reference (Mordret et al., 2018) with a better coverage for toxic species (e.g., Alexandrium and Gambierdiscus) and endosymbionts associated with coral reefs (Symbiodinium). Some genera, such as Protoperidinium, are, instead, scarcely represented and this probably explains the lack of detection of species quite common in the Gulf of Naples, such as P. diabolus and P. steinii.
In conclusion, considering the lack of reference sequences, besides the limited resolution of V4, it is clear that the dinoflagellate diversity in the Gulf of Naples is still only partially covered by the metabarcoding results, while the new findings for the area, and particularly those of species described from other oceans, will need confirmation by morphological studies. For future studies, the reference dataset should become more inclusive based on cultivation or single cell genotyping, along with long ribosomal DNA amplicons (Krehenwinkel et al., 2019) and metagenome-assembled genomes (MAGs) of species hard to cultivate (Delmont et al., 2022).
4.2 Seasonality
The analysis of the LTER–MC metabarcoding dataset shows a strong seasonal signal in the dinoflagellate assemblage of the Gulf of Naples. Samples collected over 3 years group into four main clusters broadly corresponding to the four seasons, showing each quite a wide diversity.
The similarity by seasons of the dinoflagellate associations over the 3 years considered supports the conclusions of an LM-based study that showed a remarkable stability of the whole phytoplankton assemblage occurrence at LTER-MC over three decades in spite of the high-environmental variability typical of coastal sites (Longobardi et al., 2022). Recurrent seasonal patterns of unicellular eukaryotes have also been highlighted in several multiannual studies carried out at coastal monitoring sites in the last decade. At the SOMLIT station in the eastern English Channel, cluster analysis based on OTUs' abundances indicated three main assemblages associated with spring, winter/autumn, and summer conditions (Rachik et al., 2018), while a subsequent study based on both protist abundance from LM cell counts and metabarcoding data confirms this marked seasonal pattern (Caracciolo et al., 2022). Clear recurrent patterns of several OTUs of the smallest eukaryotic fraction, mainly associated with winter and summer communities, are also detected at the long term Blanes Bay Microbial Observatory on the Mediterranean Spanish coasts (Giner et al., 2019).
The clear seasonal pattern emerging from our analysis of metabarcoding data is in apparent contrast with the view that ‘successional patterns and blooms are largely unpredictable for dinoflagellates’ (Smayda, 1997). This view may have been based mainly on species typical of the spring summer period, which generally is the season of maximum dinoflagellate abundance in temperate areas, with blooms that may vary among the years. Presumably, the view of dinoflagellates as lacking a successional pattern may also be biased by the only partial identification at species level of small and/or unarmoured species, achievable by the Utermöhl method in LM, along with the difficulty to quantify less abundant species. Both limitations are overcome by the metabarcoding approach, which allows to obtain a detailed picture of the diversity of dinoflagellates in all seasons.
The dinoflagellates dominating the metabarcoding samples of the spring and summer clusters are the ones responsible for the increase in dinoflagellate abundance observed from the onset of the stratification throughout the summer in the Gulf of Naples (Ribera d'Alcalà et al., 2004). Based on metabarcoding data, these communities are mainly characterised by chloroplast-bearing species, both unarmoured (e.g., Biecheleria, Biecheleriopsis and Gymnodinium aureolum) and armoured (e.g., Azadinium, Alexandrium, Heterocapsa, Scrippsiella and Protoceratium). The autumn association is characterized by the higher proportion of the genus Gyrodinium, including a high number of heterotrophic species (e.g., Gyrodinium helveticum).
In addition to the marked seasonal patterns, a novel result of our study is the identification of a specific winter assemblage with a high taxonomic, morphological and functional diversity, which is quite distinct from those of the other seasons in terms of overall composition and marker species. Since the winter constitutes the period of minimum dinoflagellate abundance in the annual cycle, the high-dinoflagellate diversity detected in that period could be a consequence of a lack of just a few dominant species that saturate the diversity in other seasons. However, the high number of taxa significantly associated with the winter, i.e., recurrent in that period with their highest annual abundance over the 3 years investigated, suggests the existence of a stable dinoflagellate population that is distinct from that of other seasons and well-adapted to the winter conditions. In addition to the highly diverse parasitic community, mostly only known as environmental sequences, the named species identified in winter encompass all different trophic modes, with chloroplast-bearing species mostly capable of mixotrophy (e.g., Tripos spp. and Paragymnodinium shiwaense), parasitic Amoebophrya and heterotrophic Gyrodinium. Pronounced differences in their size (e.g., the relatively small Pentapharsodinium and the large Abedinium), also point at a high-functional diversity in this community. Nonetheless, most of these species are not seen, or cannot be identified and/or quantified, in routine LM observations conducted at the LTER-MC sampling site.
The finding of a diverse winter association in relatively cold (13–16°C) and vertically mixed conditions adds a new angle to the view of dinoflagellates as always associated with stratified conditions in the warm season of the year. This view had already been challenged by the occurrence of regular or occasional blooms of specific dinoflagellates in winter (e.g., Litaker et al., 2002; Sellner et al., 1991), to which we add the detection of a non-blooming and species-rich association of dinoflagellates in winter. In the Gulf of Naples, the whole phytoplankton assemblage of this season is generally characterized by low biomass, with a high proportion of small flagellates and coccolithophores, and with dinoflagellates dominated by small-sized (<15 μm) unidentified taxa as the second group in terms of biomass. However, the possible contribution of larger dinoflagellates (Tripos spp., Abedinium dasypus, Gyrodinium fusiforme and G. helveticum) is largely underestimated by the standard counting protocols, while results of the present study show that these neglected species may constitute an important fraction of the population. Short-lasting biomass increases, mainly due to diatoms, may take place also in winter, associated with low-surface salinity in periods of calm and sunny weather that can occur even in this period of the year (Zingone et al., 2010). Both the constantly low-standing stocks and these transient pulses of autotrophic biomass may be able to sustain the heterotrophic and mixotrophic component of the dinoflagellate association that characterizes the winter period in our study area.
5 CONCLUSION
This metabarcoding study has opened a new window on dinoflagellate species diversity and ecological characteristics, with results that can be better interpreted in the frame of the background information deriving from long term observations conducted in the Gulf of Naples. The discovery of many ASVs attributed to taxa so far unreported in an intensively investigated area demonstrates that diversity assessments by means of metabarcoding may be far more effective than by means of morphological approaches. Yet there is still a need of classical studies aimed at species characterization in order to obtain a comprehensive reference database allowing for a more complete picture of dinoflagellate diversity. The discovery of a complex and distinct winter dinoflagellate community indicates that there is still much to learn about the diversity, biogeography and ecology of marine dinoflagellates, and metabarcoding can be a powerful ally in generating the new and deeper knowledge that is requested for this group as well as for the whole marine microbial compartment.
ACKNOWLEDGMENTS
The authors would like to thank Marco Cannavacciuolo, Augusto Passarelli, Ferdinando Tramontano, Gianluca Zazo and the crew of the R/V Vettoria, for sampling and data collection at sea. We also thank the Marine Research Infrastructure of the Stazione Zoologica for acquiring, processing, and managing the environmental data, and the entire LTER–MC Team for continued collaboration in the project. The authors also thank M.P. Tomasino for help with filtering and DNA extraction.
FUNDING INFORMATION
SM has been supported by a PhD fellowship funded by the Stazione Zoologica Anton Dohrn (Open University – Stazione Zoologica Anton Dohrn PhD Program); RP was supported by the project FIRB Biodiversitalia; GZ was supported by ASSEMBLE PLUS funded by the European Union's Horizon 2020 R&I Programme under grant agreement No 730984. Metabarcoding data were obtained within the projects FIRB Biodiversitalia (grant number RBAP10A2T4) and the Italian Flagship Project RITMARE, both funded by the Italian Ministry of Education, University and Research (MIUR). The LTER–MC program is funded by SZN.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interests.
Open Research
DATA AVAILABILITY STATEMENT
Raw metabarcoding reads used in this study were submitted to the European Genome-phenome Archive EBI–ENA under the Bioproject number PRJEB56637.




