Vertical and temporal distribution of chytrids infecting diatoms in the Gulf of Naples (Italy, Mediterranean Sea)
Abstract
The presence of phytoplankton parasites along the water column was explored at the Long-Term Ecological Station MareChiara (LTER-MC) in the Gulf of Naples (Mediterranean Sea) in October 2019. Microscopy analyses showed diatoms dominating the phytoplankton community in the upper layers (0–20 m). Metabarcoding data from the water column showed the presence of Chytridiomycota predominantly in the upper layers coinciding with the vertical distribution of diatoms. Laboratory incubations of natural samples enriched with different diatom cultures confirmed parasitic interactions of some of those chytrids—including members of Kappamyces—with diatom taxa. The temporal dynamics of diatoms and chytrids was also explored in a 3-year metabarcoding time-series (2011–2013) from surface waters of the study area and in sediment samples. Chytrids were recurrently present at low relative abundances, and some taxa found to infect diatoms in the incubation experiments were also identified in the ASV time-series. However, co-occurrence analyses did not show any clear or recurrent pairing patterns for chytrid and diatom taxa along the 3 years. The chytrid community in the sediments showed a clearly different species composition compared with that recorded in the water column samples, with higher diversity and relative abundance. The combination of observations, incubations, and metabarcoding confirmed that parasites are a common component of marine protist communities at LTER-MC. Host–parasite interactions must be determined and quantified to understand their role and the impact they have on phytoplankton dynamics.
1 INTRODUCTION
The role of parasitism in marine food webs has been historically overlooked, and knowledge on the diversity and ecology of parasitic organisms is still scarce (Lafferty et al., 2006). However, the interest in parasitism on phytoplankton has increased during recent years because new molecular tools suggest these interactions are more common and abundant in marine waters than previously thought (de Vargas et al., 2015; Lima-Mendez et al., 2015).
Diatoms are one of the main phytoplankton groups in marine waters (Armbrust, 2009; Falkowski, 2002; Sarthou et al., 2005) and are affected by a wide range of parasitic groups, whose diversity and prevalence are still underestimated (Scholz et al., 2016). In freshwater systems, parasites of diatoms, such as chytrid fungi (Chytridiomycota), have been largely studied because of the epidemics they frequently cause in lacustrine phytoplankton (Gleason et al., 2014; Gsell et al., 2013; Rasconi et al., 2012). Chytrids have a complex life cycle during which they can produce a high number of motile zoospores to infect new hosts, thus being called zoosporic parasites. Chytrids can be saprotrophs, obligate parasites, or facultative parasites, meaning that they can grow under diverse trophic lifestyles depending on the host and organic matter availability (Frenken et al., 2017). Likewise, some chytrid species seem to be highly specific in their interactions, while others show a generalist behavior, probably forming a continuum between the two strategies (Kagami et al., 2021; Van den Wyngaert et al., 2018). In any case, little is known about the specificity of their interactions, but they have been shown to play an important role in the control of phytoplankton population size and seasonal succession (Ibelings et al., 2011), as well as in the transfer of matter from inedible substrate to consumers (Kagami et al., 2014; Klawonn et al., 2021).
In contrast to freshwater environments, chytrids remain poorly characterized and understood in the marine ecosystem. Difficulties to detect and identify the causative organisms have so far hampered the inclusion of parasitism in current marine ecology and biogeochemical models, but recent advances in molecular techniques and the combination with microscopy observations have provided evidence of their wide occurrence in marine habitats. Several diatom–parasite interactions in the sea have been recently described including those involving vampyrellids (Alves-de-Souza et al., 2019), Oomycetes (Garvetto et al., 2018), and chytrids (Garvetto et al., 2019), but the knowledge on parasites diversity is still scarce, concerning only a few host–parasite systems, often with incomplete taxonomic characterizations. Thus, species-specificity of interactions is still insufficiently understood (Frenken et al., 2017; Ilicic & Grossart, 2022; Scholz et al., 2016). Therefore, investigations are being carried out to determine the diversity and occurrence of different groups of marine zoosporic parasites infecting phytoplankton, mostly focusing on fungal chytrids, in environments with freshwater-marine gradients such as Artic or Antarctic waters (Cleary & Durbin, 2016; Hassett & Gradinger, 2016; Kilias et al., 2020). Likewise, the presence of parasitic organisms is being explored in available temporal series (Christaki et al., 2017; Gutiérrez et al., 2016; Käse et al., 2021). Some studies have also focused on determining the presence of zoosporic parasites in different sites of the Mediterranean Sea, mostly represented by coastal environments, some of them under phytoplankton bloom conditions, and including information from sediments (Chambouvet et al., 2014; Fernandez-Valero et al., 2022; Reñé et al., 2021; Richards et al., 2015; Siano et al., 2011). In any case, available information in marine waters is still scarce and particularly in the Mediterranean Sea.
Different parasitic groups show a heterogeneous distribution along the water column in freshwater and marine systems (Gsell et al., 2013; Siano et al., 2011; Xu et al., 2018), suggesting that the distribution of each parasitic group is not random in the water column and depends on many abiotic and biotic factors such as light availability, water column structure, host distribution and abundance, and parasite motility. These factors not only affect the distribution but also affect the prevalence of parasites. For example, high irradiances increased cyanobacteria growth and parasite transmission in a chytrid-cyanobacterium system (Tao et al., 2020), while high temperatures inhibited chytrid infections on a cyanobacterial host, indicating a possible thermal refuge (McKindles et al., 2021). By contrast, low temperature decreased infections in a Parvilucifera (Perkinsea)-dinoflagellate system (Schmitt et al., 2022).
In the Gulf of Naples (Tyrrhenian Sea), research has been performed on plankton ecology and taxonomy especially over the last three decades (see Zingone et al., 2019 for a review), addressing interspecific relationships, such as food webs (D'Alelio et al., 2015), parasitic infections in copepods (Ianora et al., 1987, 1990), or viral infections in phytoplankton (Zingone et al., 1999), among other ecological mechanisms. Preliminary HTS metabarcoding studies including samples from the Gulf of Naples revealed that chytrids are an important component (up to 61% of eDNA reads) of marine fungi (Richards et al., 2015). However, information is missing on their vertical distribution, and their relationships with the planktonic organisms have never been explored in the study area.
Diatoms are the main phytoplankton group in the Gulf of Naples, where they are responsible for the recurrent seasonal biomass increases from spring through autumn (Ribera d'Alcalà et al., 2004). It is therefore relevant to investigate parasites that could regulate their abundance and seasonal cycles in the area. This study focuses on the occurrence and recurrence of diatom parasites in the Gulf of Naples, with two main objectives. First, we aimed at the identification of specific diatoms–parasite interactions and their distribution along the water column, based on ad hoc sampling and incubation experiments conducted in October 2019, complemented by morphological and metabarcoding data analyses. Additionally, the presence of parasitic organisms was explored in a 3-year metabarcoding time-series from the LTER-MareChiara (LTER-MC) site in order to identify potential host–parasite co-occurrences and seasonal dynamics.
2 MATERIALS AND METHODS
2.1 Sampling and processing of seawater
The water column was sampled on 8th, 15th, and 22nd October 2019 (MC1347, MC1348, and MC1349) at the LTER-MC sampling site, which is located 2 miles offshore the city of Naples, on the 75 m isobath, at the border between the coastal eutrophic system influenced by land runoff and the offshore oligotrophic waters with characteristics of the southern Tyrrhenian Sea (Mediterranean Sea). Water temperature, salinity and fluorescence profiles were acquired by means of SBE 911plus CTD. The CTD was connected to a Sea-Bird Electronics automatic Carousel sampler. Data were processed (mediated to 1 m) using the SeaSoft Data Processing Software. Surface waters correspond to 0.5 m depth. However, they are reported as 0 m across the study for clarity. Samples for dissolved inorganic nutrient analyses (nitrites, nitrates, silicates, and phosphates) were collected from the Niskin bottles at 10 depths (0, 2, 5, 10, 20, 30, 40, 50, 60 and 70 m) and immediately stored in 20-ml high-density polyethylene vials at −20°C until the analysis. The analyses were carried out with a five-channel continuous flow autoanalyzer (Flow-Sys System). Additional water samples were filtered on Whatman glass-fiber filters to determine chlorophyll-a concentration. The filters were immediately stored in liquid nitrogen, and the analyses were performed using a SHIMADZU (mod. RF-5301PC) spectrofluorometer. Details of the methods are found in Sabia et al. (2019).
Additional seawater from selected depths—surface (0 m), 5, 10, 20, 40, and 60 m—was obtained from a second Rosette cast using 10-L Niskin bottles. Subsamples were collected in 250-ml glass bottles and immediately fixed with Lugol's iodine for the phytoplankton community analysis. The remaining seawater was pre-filtered in situ using a 200 μm net-mesh and an appropriate volume (2–4 L) of seawater sample from each depth was filtered onto 0.8 μm 47 mm polycarbonate filters (DHI) using a vacuum pump at low pressure. One liter of seawater from each depth was collected in polycarbonate bottles and brought to the laboratory for further experiments (see below). The benthic compartment harbors its own communities, but also act as reservoirs of planktonic species and accumulates organisms sinking from upper layers. Thus, sediment samples were obtained during the second sampling using a gravity sediment corer deployed at approx. 75 m depth. Around 20 g of sediment, corresponding to the upper centimeters, was separated and placed in a collection tube. The sediment and the filters were immediately frozen at −80°C until processed for DNA extraction.
2.2 Incubations with cultured diatoms
Back in the labortory, 1 L of each sample was concentrated using a 10 μm mesh net, and an approximate volume of 5 ml was transferred to six-wells culture plate. Parasitic infections are easily propagated when phytoplankton populations reach high abundances. Therefore, each well was enriched with additional 5 ml of a different healthy clonal culture of diatoms (Table S1) to promote infections on those potential hosts, resulting in the incubation of six different wells for each sample. Those well-plates were regularly examined with an inverted light microscope Leica DMIL LED along 1 week to detect infected diatoms. Cells apparently infected were photographed using a Leica MC170 HD digital camera, and the development of the infections was observed over the days to determine the parasites' life-cycle. After 2 weeks, the total volume of the six wells corresponding to each depth and sampling date was mixed (approx. 60 ml), filtered onto 0.8 μm 47 mm polycarbonate filters (DHI) using a vacuum pump at low pressure and immediately frozen at −80°C until DNA extraction. Individual infected diatom cells were isolated when observed in any of the incubations performed, and transferred to new wells, adding the appropriate cultured host cells, to establish monospecific parasite–host co-cultures. Unfortunately, the cultures did not survive, impeding a further morphological and molecular characterization.
2.3 Phytoplankton
Variable volumes of seawater (10–50 ml) were settled depending on cell concentration. For each sample, one or two transepts were counted representing ca 1/60 and 1/30, respectively, of the whole bottom area of the sedimentation chamber. Identification and counts of specimens were performed according to the Utermöhl method (Edler & Elbrächter, 2010) using an inverted light microscope Zeiss Axiovert 200 (Carl Zeiss) at 400× magnification. Specimens were identified at the lowest taxonomic level possible according to Tomas (1997) and following taxonomic changes in the most relevant and updated literature.
2.4 Metabarcoding analyses
Total genomic DNA was extracted from all filters, previously cut in small pieces, using the DNeasy Blood & Tissue kit (Qiagen) as follows. A first cell lysis step was performed embedding the filter pieces in 200 μl PBS, 20 μl Proteinase K, and 200 μl Buffer AL and incubating them at 56°C for 10 min. Successive steps were conducted according to the manufacturer's instructions. Two replicates of 5 g were collected from the sediment sample, and total genomic DNA was extracted independently for each replicate using the DNeasy PowerMax soil kit (Qiagen) following the manufacturer's instructions. DNA was eluted in 5 ml and concentrated to a final volume of 200 μl using Amicon Ultra centrifugal filters. Genomic DNA samples were sent to an external sequencing service (AllGenetics), where a first PCR was conducted with eukaryotic universal primers TAReukFWD1 and TAReukREV3 to amplify the V4 region of 18S rRNA gene, followed by MID-indexing of the products, their pooling in equimolar amounts and sequencing in a MiSeq PE300 Illumina run. The raw reads obtained were trimmed from the adaptors using cutadapt v.1.16 (Martin, 2011). The DADA2 pipeline (Callahan et al., 2016) was used to filter, dereplicate, and trim low-quality sequences (maxEE = 4, 6; truncLen = 280, 270), merge the paired ends (minOverlap = 10) and remove chimeras. The resulting ASVs (amplicon sequence variants) were classified using vsearch v2.8.1 global alignment algorithm (Rognes et al., 2016), against the PR2 v4 database (Guillou et al., 2013); the identity cutoff was 90%. Sequence analyses were run at the Marine Bioinformatics Service (MARBITS) of the Institut de Ciències del Mar (ICM-CSIC) in Barcelona. A summary of raw reads, final amplicons, and number of ASVs obtained for each sample is shown in Table S2. The metabarcoding data were analyzed using the package Phyloseq (McMurdie & Holmes, 2013), and all graphs were constructed using ggplot2 (Wickham, 2016). Two samples with <100 reads were removed, corresponding to the natural samples from 8th October 0 m and 22nd October 5 m. The ASVs belonging to metazoans, and the 8 ASVs belonging to cultured diatoms used to enrich the incubations, were removed from the dataset. The richness and diversity of samples were evaluated by Chao1 and Shannon indices. Barplots were constructed using the data normalized by sample relative abundance. Non-metric multidimensional scaling (NMDS) using Bray–Curtis distances was performed to analyze the dissimilarity between sampling dates and depths using the data normalized by sample relative abundance.
The presence of chytrids and potential chytrid-diatom co-occurrences was explored in a V4-18S rDNA metabarcoding dataset generated through Illumina HTS from environmental DNA extracted from plankton samples collected in surface waters at LTER-MC on 48 dates over 3 years from January 2011 to December 2013 (Gaonkar et al., 2020). For details of sampling, DNA processing and Illumina high-throughput sequencing, see (Piredda, Tomasino, et al., 2017). One sediment sample obtained at 75 m on 7th February 2014 at LTER-MC and treated as described in Piredda, Sarno, et al., (2017) was also included. ASVs were generated as follows: quality metrics of demultiplexed reads were first assessed using FastQC. Reads were then trimmed with cutadapt (Martin, 2011) following a two-stage procedure: in the first round, 5′ primers were trimmed, discarding reads if the primer was not found; in the second round 3′ primers were trimmed, but keeping the reads without primers. In both rounds adapter mismatch was set at 0.25 and terminal Ns removed if present. The 2-stage procedure avoids discarding read pairs from amplicons sized between 250 and 490 bp, long enough for the reads to overlap but where 3′ primers are truncated or absent. Contig assembly, denoising, dereplication, and chimera screening were performed with the DADA2 R library (Callahan et al., 2016), using the default parameters described in the MiSeq tutorial, except for allowing up to six mismatches in read merging (function mergePairs, parameter: maxMismatch = 6). Resulting ASVs were then classified with BLAST (Altschul et al., 1997) using an identity cutoff of 90% against the PR2 v4.12 reference database (Guillou et al., 2013). Raw metagenomic reads corresponding to October 2019 and 2011–2013 samplings were submitted to the NCBI Sequence Read Archive under the Bioproject number PRJNA878459 and to the European Genome-phenome Archive EBI-ENA under the Bioproject number PRJEB56637 respectively.
The ASV assigned to chytrids obtained during the sampling campaign in 2019 and those from the metabarcoding dataset of LTER-MC (2011–2013) were extracted. The correspondence of ASVs from the two datasets was evaluated by the identity percentage obtained using BLAST, only considering those with an identity cutoff of 99% and 90% coverage. The pairwise-correlations between diatoms and chytrids were computed based on relative abundance matrices calculated for the natural samples of vertical profiles and for the time-series samples using the function rcorr from Hmisc package. The correlation coefficient cutoff was 0.8 and p < .05 in both cases. For these analyses, we only used the ASVs present in more than one sample. The pairwise networks were plotted using Cytoscape v.3.9.1 (Shannon et al., 2003). A schematic figure summarizing the different approaches conducted and their combination can be found in Figure S1.
3 RESULTS
3.1 Physical and chemical conditions during the sampling campaign
The three CTD profiles performed concurrently with the biological sampling showed a clear thermocline between 20 and 30 m depth (Figure S2). The upper layer was predominantly mixed showing homogeneous values in terms of water temperature (approx. 24°C) and salinity (38.2). Fluorescence peaks were observed at 5–30 m, depending on the sampling day. Secchi depth ranged between 10 and 15 m. The water column below the thermocline showed a gradual decay in temperature and fluorescence, reaching values around 14°C and 0 RFU. Inorganic nutrients showed higher values in deeper layers (Figure S3), especially NO3 and NO2, which showed concentrations <0.5 and <0.2 mM in layers above 20 m, respectively, increasing from 30 to 40 m downwards to maximum values of 2.3 and 0.5 mM. Maximum SiO2 concentrations >2 mM were also detected in deeper layers, but a decrease from the surface to 10 m depth was observed on 8th and 15th October, followed by a downward increase of their concentrations. PO4 concentrations were low, with a maximum of 0.12 mM at 5 m (15th October), and did not show remarkable differences along the water column. Chlorophyll-a concentrations were higher in upper layers between 0 and 10 m, ranging between 0.4 and 1.3 mg m-3 and gradually decreased in deeper layers, especially evident on 15th October. Maximum concentrations on the three sampling dates were 0.9, 1.3 and 0.5 mg m-3, respectively.
3.2 Microbial community composition
Light microscopy observations (Figure 1) showed diatoms dominating the phytoplankton communities at 0, 5 and 10 m on 8th and 15th October, reaching maximum abundances of 4 × 106 cells L-1 at surface on 15th October. Diatoms gradually became less abundant along the water column, being almost absent at 40 and 60 m. The same trend was observed on the third sampling date, when diatom abundance was lower (6 × 105 cells L-1 at surface). Diatoms represented >40%–70% of cell counts in all samples from 0 to 20 m, except on 22nd October, when their relative abundance represented 25%–45% of total cell counts in the 0–20 m samples. In all cases, diatoms represented <20% of cell counts in 40 and 60 m samples. Small-sized flagellates (<10 μm) were the second major group and largely dominated the community in terms of abundance at 40 and 60 m. Dinoflagellates were also present in the samples, but always at low abundances (<105 cells L-1), representing <5% of cell counts from 0 to 20 m.
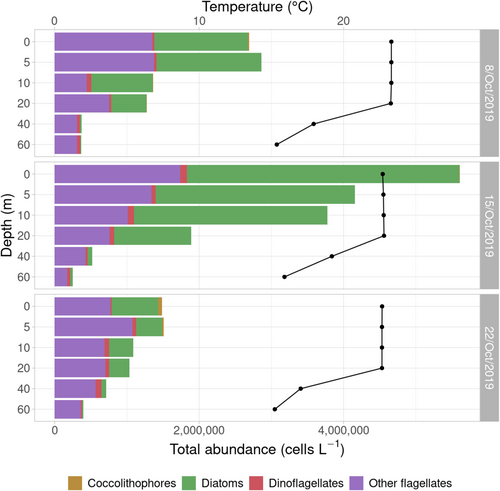
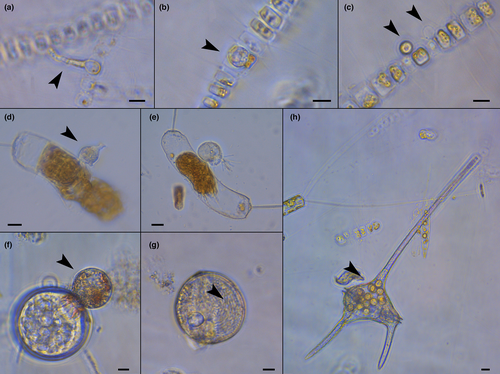
In the diatom community, Leptocylindrus aporus was predominant at 0–20 m, while its abundance decreased at 40 and 60 m, where other centric diatoms became more abundant. On 22nd October, L. aporus proportion remained constant along the water column, and differences in composition were mostly due to the presence of Chaetoceros spp. in upper layers, mainly C. tenuissimus (data not shown). The microscopy analyses of the natural samples also allowed the observation of diatom cells presumably infected by parasitic organisms (Figure 2a–f), as well as other infected phytoplankton members like dinoflagellates (Figure 2g,h).
Compared with the microscopy results, metabarcoding analyses of the protist community showed a different group percentage composition (Figure 3), with Dinoflagellata, that is, dinoflagellates (Dinophyceae) and Syndiniales, representing >75% of reads retrieved. This percentage slightly decreased in deeper layers (40 and 60 m), where Dinoflagellata represented around 60% of the reads and diatoms (Bacillariophyta) and other minor groups showed a larger relative abundance compared to upper layers. In terms of major groups, the community composition at each depth was highly similar among the three samplings dates.
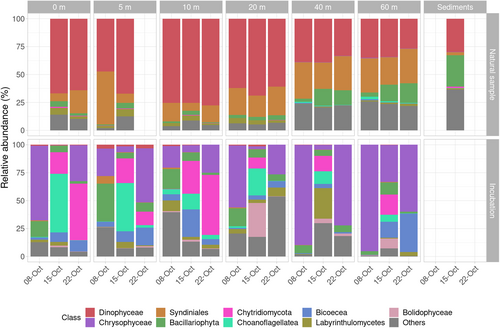
The differences in community structure and composition observed for samples above and below the thermocline were also reflected in the community dissimilarity analysis, showing a clear separation between them (Figure S4). Some groups, including phytoplankton parasites such as Chytridiomycota, Oomycota, Pirsonia or Perkinsea, were present in the community but, except for chytrids with relative abundances spanning from 0% to 1.1%, they always represented abundances <0.1%, and were grouped as “Others” in Figure 3.
The sediment sample showed a similar composition to seawater samples in terms of main taxonomic groups. However, Syndiniales were almost absent, the percentage of Dinophyceae resembled that obtained at 40 and 60 m, and the contribution of dinoflagellates and diatoms was comparable, representing 30% of reads each (Figure 3). The presence and contribution of other less abundant groups were similar to those of the water samples from 40 and 60 m. However, Chytridiomycota were present in higher abundances than in the water column, representing 0.8% and 1.6% in both replicates. The similarity in the community from 40, 60 m and sediments was reflected in alfa-diversity analyses, showing higher richness (Chao1 index) and diversity (Shannon index) values for these samples from deeper layers (Figure S5).
3.3 Detection of parasitic infections in incubation experiments
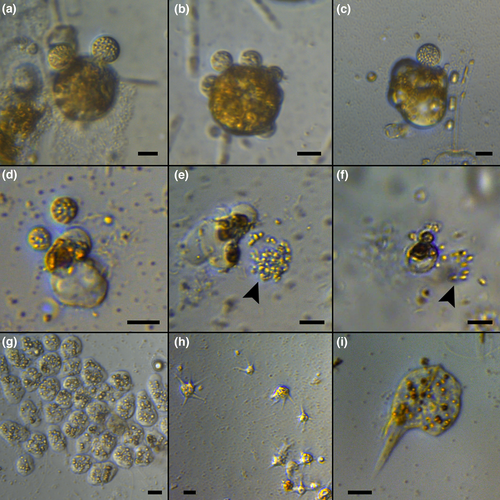
Regular observations of culture plates with concentrated natural samples enriched with diatom cultures were performed for 1 week. At the end of the incubation period, the presence of cells infected by chytrids was confirmed for the 15th and 22nd October samples, where chytrid sporangia were observed in incubations enriched with Ditylum brightwellii (Figure 4a–c) and Chaetoceros affinis (Figure 4d). The release of zoospores was observed during light microscopy observations (Figure 4e,f). Chytrid infections were observed in most samples corresponding to upper layers (0 and 20 m). Efforts to maintain those chytrids in culture were unsuccessful and they were finally lost, preventing their characterization and identification at the species level. Besides chytrids, other microorganisms increased their abundance, in some cases actively feeding on diatoms present in the well (Figure 4g–i).
Metabarcoding results obtained after enriching the natural samples with selected diatom cultures and incubating them for 1 week allowed to determine the organisms that encountered optimal conditions for their growth (Figure 3). The number of reads obtained for incubated samples was in the same range as those from natural samples (50,000–90,000), but the number of ASVs was much lower (Table S2). While ASVs in natural samples ranged between 70 and 750 per sample, their number in incubations samples ranged between 25 and 200. Chytridiomycota (chytrids) were present in all samples, with abundances up to 50% of reads in some of them, like at 0 and 10 m on 22nd October. The samples from 8th and 22nd October were also dominated by other organisms such as the non-photosynthetic bacterivore Spumella sp. (Chrysophyceae), while the diatom-attached bacterivore Salpingoeca sp. (Choanoflagellates) was detected in incubations from 15th October. Labyrinthulomycetes representatives including diatom detritus feeders were also detected in some incubations.
Given the dominance of chytrids in the incubations and the fact that they included parasitic organisms of diatoms, a detailed analysis of chytrids was performed (Figure 5). A total of 34 different ASVs belonging to chytrids were recorded in the October dataset, including 16 samples from natural seawater, 18 from incubations, and one from sediments, which was not used in incubation experiments. A total of 13 ASVs were exclusively detected in the sediment sample, which showed a higher chytrid diversity than seawater samples, while only two ASVs (ASV11 and ASV511) were detected in both seawater and sediment samples. The 19 remaining ASVs were exclusively detected in seawater samples. Four of them (ASV11, ASV60, ASV1527, and ASV2804) were detected in both natural samples and incubation experiments, and 10 were exclusively detected in incubations. The relative abundance of chytrids was always low in the natural samples, but increased in the incubation experiments. Chytrids were detected at 5, 10, and 60 m in the natural samples of 8th October (surface sample data are missing), and in the incubated samples of the same date from 5, 10, 20, and 40 m. The diversity of chytrids was high, but they always represented low relative abundances (<0.1%). In the 8th October 5 and 10 m samples, the ASVs detected in nature and after the incubations were different. By contrast, the same ASVs were detected in nature and incubations of profiles from 15th and 22nd October. In both cases, only two chytrid ASVs were detected after the incubation period, mainly ASV11 and ASV60 at lower abundances, thus representing low diversity. However, they showed high relative abundance in the incubations, in some cases up to 50% of the reads. On 15th October, ASV11 was detected in nature from the surface to 20 m. In the incubations, this ASV was detected at all depths, even though its relative abundance was lower from 20 to 60 m. On 22nd October, ASV11 was detected at surface, 10 and 20 m in nature (5 m data are missing), while ASV11 and ASV60 were detected at surface, 5 and 10 m in incubations. The presence of chytrids was not detected from 20 m downwards. Positive co-occurrence relationships with diatoms were found for two chytrid ASVs, ASV11 and ASV2813 (Figure S6). The former was present in both nature and incubations, whereas ASV2813 was only found in nature. ASV11 co-occurred with three different ASVs assigned to Thalassiosira sp., while ASV2813 also co-occurred with one of the Thalassiosira sp. ASV. A similarity search of ASV11 sequence in NCBI database showed >90% similarity with several environmental sequences belonging to the Rhizophydiales order, and a maximum similarity of 99.7% with sequence LN827831, corresponding to an environmental sequence of Kappamyces sp. obtained from the Gulf of Naples in 2009.
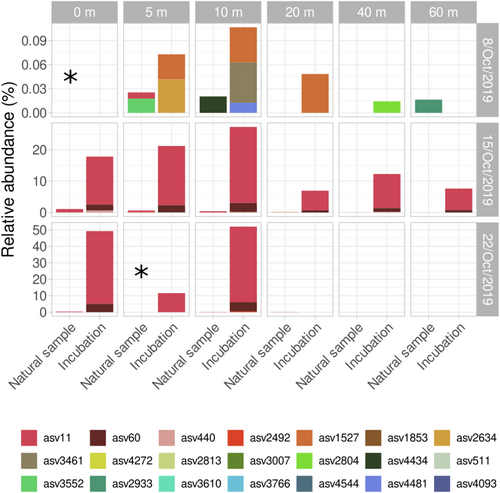
3.4 Diversity and temporal distribution of chytrids and their association with diatoms in the Gulf of Naples
Given the detection of chytrids during the sampling campaign performed, and the confirmation of their real interaction with diatoms, the presence of chytrids was explored in a metabarcoding dataset from LTER-MC, consisting of surface water data obtained on 48 sampling dates from 2011 to 2013 (Figure 6). A total of 238 ASVs classified as Chytridiomycota were identified in the time series, and they were consistently present over the 3 years, commonly representing <0.5% of the protist community, but showing peaks on June 2011, December 2011, November 2012, and December 2013, with maximum relative abundance of 1.3% in November 2012. Those peaks did not show a relationship with high relative abundances of diatoms. A total of 46 ASVs were detected in the sediment sample analyzed, representing an abundance of 1.1% of the total protist community. Forty of them were exclusively detected in the sediment sample, and only six were also detected in surface waters.
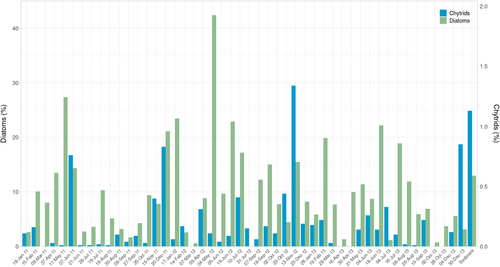
Among the chytrid ASVs from the sampling campaign of October 2019, 16 showed identity >99% to 25 ASVs detected during the time-series (Table 1), including ASV11 and ASV60, which represented the chytrid ASVs predominantly detected during the incubation experiments. These two ASVs and ASV1853 only differed by a single nucleotide among them and were treated as belonging to the same chytrid species in following analysis. In the time-series, the five ASVs showing >99% identity with ASV11–ASV60–ASV1853 (Table 1) appeared in 16 of 48 surface samples, records being scattered along time and showing no seasonal pattern or correspondence with high diatom abundances. Among them, pairwise correlations only showed a positive relationship between the chytrid ASV4881 and the diatom ASV7687, the latter corresponding to the centric genus Actinocyclus, Coscinodiscophyceae (r = .86, p < .01). As previously described, some ASVs were exclusively detected in the sediment samples analyzed. Seven of the 15 ASVs obtained in sediments processed during the sampling campaign of October 2019 showed >99% identity with ASVs present in the sediment sample available from February 2014.
| October 2019 | Reads | % identity | Time-series | Reads |
|---|---|---|---|---|
| ASV11 | 40,742 | 100 | ASV1193 | 1307 |
| ASV11 | 99.7 | ASV4881 | 2 | |
| ASV11 | 99.7 | ASV26849 | 113 | |
| ASV11 | 99.5 | ASV2796 | 339 | |
| ASV11 | 99.2 | ASV17747 | 7 | |
| ASV60 | 4702 | 99.7 | ASV1193 | |
| ASV60 | 99.5 | ASV4881 | ||
| ASV60 | 99.5 | ASV26849 | ||
| ASV60 | 99.2 | ASV2796 | ||
| ASV1853 | 44 | 99.7 | ASV1193 | |
| ASV1853 | 99.5 | ASV4881 | ||
| ASV1853 | 99.5 | ASV26849 | ||
| ASV1853 | 99.2 | ASV2796 | ||
| ASV440 | 393 | 100 | ASV5756 | 79 |
| ASV511 | 317 | 100 | ASV2835 | 329 |
| ASV884 | 140 | 100 | ASV2489 | 417 |
| ASV1378 | 73 | 99.7 | ASV7407 | 48 |
| ASV2166 | 32 | 100 | ASV829 | 2120 |
| ASV2166 | 99.7 | ASV11970 | 17 | |
| ASV2492 | 23 | 99.2 | ASV11330 | 19 |
| ASV2499 | 23 | 100 | ASV6991 | 54 |
| ASV2813 | 16 | 100 | ASV2011 | 601 |
| ASV2813 | 99.2 | ASV4781 | 118 | |
| ASV2933 | 14 | 100 | ASV1297 | 1165 |
| ASV2933 | 99.7 | ASV20796 | 5 | |
| ASV2933 | 99.5 | ASV7209 | 50 | |
| ASV2933 | 99.2 | ASV13331 | 13 | |
| ASV3183 | 11 | 100 | ASV2019 | 596 |
| ASV3552 | 7 | 100 | ASV4784 | 118 |
| ASV3766 | 6 | 100 | ASV741 | 2522 |
| ASV3766 | 99.7 | ASV7435 | 47 | |
| ASV3766 | 99.2 | ASV6718 | 58 | |
| ASV4272 | 3 | 100 | ASV6718 | 58 |
| ASV4272 | 99.2 | ASV741 | 2522 |
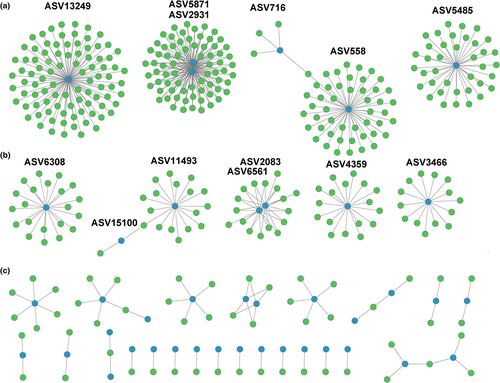
Pairwise correlations were also computed for all chytrid-diatom co-occurrences. Positive relationships were found for 41 chytrid ASVs, showing correlations with 392 different diatom ASVs (Figure 7, Table S3). However, different patterns were observed. Some chytrids ASVs, that is, ASV13249, ASV2931, ASV4871, ASV558, and ASV5485, showed positive correlations with numerous ASVs assigned to diatoms, ranging between 92 and 38 (Figure 7a). Six more chytrid ASVs showed significant co-occurrences with some diatom ASVs, ranging between 21 and 14 simultaneous positive correlations (Figure 7b). Finally, numerous ones only showed a positive relationship with a limited number of diatom ASVs, ranging from six to a single one (Figure 7c). The taxonomic resolution of V4 18S rDNA did not allow a robust taxonomic assignment of chytrid ASVs, which remained classified as uncertain Chytridiomycetes or as Spizellomycetales/Rhizophlyctidales. They showed positive relationships with diatoms of diverging affiliation, including representatives of Thalassiosira spp., Chaetoceros spp., Rhizosolenia delicatula, Skeletonema spp., Mediophyceae, and pennate diatoms such as Nitzschia spp. or Navicula spp. Thus, a taxonomic specificity between chytrids and hosts was not apparent.
4 DISCUSSION
In this study, the interactions among some chytrids and their hosts were unravelled thanks to the combination of microscopy, molecular, and cultivation approaches. Such approach has been used in different recent studies, for example, Van den Wyngaert et al. (2022) and Fernandez-Valero et al. (2022). Molecular approaches allow the detection and identification of chytrids, not straightforward using traditional microscopy techniques. By contrast, many aspects such as parasite–host interactions, and ecological or morphological features cannot be determined unless efforts are conducted to observe the samples by microscopy and cellular cultures are established. Regarding the metabarcoding approach, other markers such as 28S rDNA or ITS are widely used for fungi, providing a better taxonomic resolution. However, data available for the time series were V4 18S rDNA. Thus, information from the same genetic marker was generated in this study to ensure the possibility of comparing homogeneous data.
The temporal patterns of those interactions in the coastal waters of the Gulf of Naples indicate that, in nature, chytrids generally infect phytoplankton representatives at low prevalence but, under certain conditions still to be clarified, they may propagate and increase their impact on those populations. In the vertical profiles, chytrids were predominantly detected in the upper layers of the water column, matching the vertical distribution of diatoms, which appeared to be mostly present above the thermocline. Diatoms dominated in the well illuminated upper layers, while their abundance was much lower from 20 m downwards. This vertical distribution, along with a secondary diatom bloom, is commonly observed in the Gulf of Naples during stable weather conditions in autumn (Ribera d'Alcalà et al., 2004; Zingone et al., 1995).
The structure of the protist community determined by metabarcoding was markedly different from that observed by microscopy. Alveolata representatives, that is, Dinophyceae and Syndiniales, dominated at all depths, in agreement with the results of many metabarcoding studies highlighting the importance of these groups in marine waters (de Vargas et al., 2015; Pernice et al., 2013; Piredda, Tomasino, et al., 2017). However, it is well-known that Alveolata have large genomes with numerous copies of ribosomal genes (Medinger et al., 2010) and, consequently, the high relative abundance of reads likely represents an overestimation of the actual cell abundance in natural communities due to possible methodological artifacts.
Given the compositional nature of metabarcoding data, the wide dominance of Alveolata sequences hampers an adequate coverage of less abundant organisms, whose characterization would require the use of selective, target-specific primers. A maximum chytrid relative abundance of 1.1% was recorded at 0 m on 15th October, but the relative abundance of chytrids was consistently <0.5% across the samples explored in the vertical profiles. Similar relative abundances were seen in the historic time series for the studied station, in which only four peaks were observed and always with relative abundance <1.5%. These results are in agreement with fungal diversity and abundance data obtained from publicly available metabarcoding datasets, which showed a mean global relative abundance of chytrids of 1.3% of eukaryotic sequences, representing 36% of the fungal community (Hassett et al., 2020). However, a higher contribution of chytrids was found in environments with salinity values differing from the global mean, such as the sea ice, Baltic Sea, or the Red Sea (Hassett et al., 2020). Several studies showed a wide variation in chytrid abundances (Banos et al., 2020; Comeau et al., 2016; Hassett et al., 2017; Taylor & Cunliffe, 2016). Those variations were related to equally varying abundances of potential hosts in the community, but a higher contribution of parasitic chytrids has also been observed nearshore compared to the open sea (Rubin-Blum et al., 2022). The low chytrid contribution despite high diatom abundance detected during this study could be related to the hydrographic conditions of the Gulf of Naples, where frequent exchanges occur with oligotrophic Tyrrhenian waters. The resulting dilution effects could prevent the accumulation of parasitic interactions and thus the increase of their prevalence.
The low abundance of chytrids prevented their observation in natural samples by microscopy, but the laboratory incubations adding diatoms as potential hosts showed that parasites, mainly fungal chytrids, were active and able to infect diatoms in the natural environment of the Gulf of Naples, prompting further explorations through the analysis of metabarcoding data on both natural samples and diatom-enriched experiments. In the former, the low chytrid abundance was probably close to the detection limit of the sequencing depth used in this study. Indeed, there were cases of chytrid sequences that were absent from the natural sample dataset but present in the sequence data obtained from the incubation experiments. This implies that they were present at extremely low concentrations in the initial inoculum, while the incubations, enriched with high abundance of potential hosts, increased contact rates and thus their propagation and also the probability of detection.
By contrast, some ASVs present in natural samples were not detected after the diatom incubations performed on the samples of 8th October. Chytridiomycota not only infect diatoms in marine environments, but also infect dinoflagellates (Fernandez-Valero et al., 2022; Karpov et al., 2021; Lepelletier et al., 2014; Reñé et al., under review), and can also have a saprophytic behavior (Gleason et al., 2008). Therefore, the ASVs present in nature and not detected after the diatom incubations may correspond to parasites not able to infect diatoms, or at least the diatom species added in the experiment, leading to a low or null propagation of infections and hence an absence of ASVs in the incubations.
In the 8th October sampling, chytrids presented low relative abundances in both natural and incubation samples and showed a distinct species composition compared to the other two sampling dates. By contrast, the chytrids relative abundance increased in the next sampling date in line with the diatom abundance increase, and were mostly detected in the upper layers (surface to 10 m) on 15th and 22nd October, in agreement with higher diatom abundances in those layers, and mainly comprising two different ASVs. The presence of chytrids, albeit at lower abundance, in incubations from deeper layers from 15th October could correspond to the sinking of infected diatoms or to free-living infective zoospores. The dominant ASVs, ASV11 and ASV60, probably corresponding to the same species, are parasites of diatoms because those infections were confirmed by direct microscopical observations. The co-occurrence analysis showed positive relationships with some diatoms present in the natural community, such as Thalassiosira spp., suggesting these species are among the hosts they infect in nature.
The high relative abundance of chytrids in sediment samples compared with most samples from the water column, and their completely different composition, could be related to the different diatom communities, with the predominance of benthic species in sediments, and also to the higher possibility of interactions in surface sediments. In addition, their higher richness could as well be due to a contribution of saprophytic taxa that could thrive on sinking organisms. At the same time, chytrids in the sediments could represent dormant stages, constituting a reservoir of parasites that under favorable conditions could act as inoculum for recurring infections (Ibelings et al., 2004). In fact, several studies revealed the importance of chytrids in the composition of the benthic fungal community (Fernandez-Valero et al., 2022; Richards et al., 2015; Wang et al., 2017, 2018), highlighting that sediments play an important role in chytrid dynamics and suggesting a plankton-benthos coupling.
The analyses of the metabarcoding time-series from surface waters of the Gulf of Naples revealed a consistent occurrence of chytrids, showing a high diversity, with 238 different ASVs detected, albeit at low abundance. The analyzed sediments sample shows a higher diversity of chytrid ASVs, being completely distinct from those found in the surface layer. A large number of ASVs exclusive from sediments were retrieved in both sediment samples from February 2014 and October 2019, suggesting again that they correspond to species adapted to this benthic compartment and that the species composition is more stable than in the water column. Surface water chytrids did not show a seasonal pattern, but some peaks of abundance were detected along the period studied (2011–2013). These observations agree with temporal studies conducted in several other marine coastal locations, where chytrids were recurrently detected at low relative abundances but showed sporadic high abundance peaks. In some cases, the increase in chytrids showed a correspondence with increases of diatom abundances (Taylor & Cunliffe, 2016), also confirmed by microscopy observations (Gutiérrez et al., 2016; Hassett et al., 2017; Hassett & Gradinger, 2016; Kilias et al., 2020). In other cases, no relationship with any phytoplankton species was found (Christaki et al., 2017; Käse et al., 2021), illustrating that the identification of real host-parasite pairs is not easily achieved based on molecular surveys and co-occurrence analysis alone, requiring complementary approaches, including culturing and/or microscopy observations, for confirmation. In our study of the temporal series, peaks of chytrid abundance showed no relationship with increases of diatom abundances, at least in surface waters. However, not all chytrids are diatom parasites; they can be saprophytes or infect other organisms (Gleason et al., 2014). In addition, previous spatiotemporal studies highlighted that, besides being more abundant in coastal than in pelagic waters, fungi and chytrids sometimes increase their abundances with depth (Gao et al., 2010; Wang et al., 2018). Thus, information about the whole water column could also be necessary to spot these relationships in time-series analyses.
Two specific ASVs (ASV11 and ASV60) were detected in all three metabarcoding datasets: in the vertical profiles including sediments, in the incubation experiments, where the interaction with diatoms was confirmed by microscopy, and in the time series. Consequently, these ASVs correspond to parasites of diatoms with a high degree of certainty. Actually, these two ASVs and ASV1853 only differed by a single nucleotide among them. Although the evolutionary rate of the species is not known, they could be considered as different ribotypes of the same chytrid species, as was also observed for other marine chytrid species infecting dinoflagellates such as Dinomyces arenysensis (Fernandez-Valero et al., 2022). These ASVs, mainly ASV11, showed a positive correlation with occurrences of Thalassiosira sp. in the natural communities sampled in this study, suggesting this specific host–parasite interaction occurs in nature. As mentioned, ASV11 was present in the time-series but only a weakly supported relationship was observed with co-occurring diatoms, specifically with Actinocyclus sp. Based on sequence identity and taxonomic classification of closest relatives, this ASV corresponds to a Kappamyces (Rhizophydiales) representative. The members of this genus are described as freshwater species growing as saprotrophs on pine pollen (Letcher & Powell, 2005). Its detection in previous sampling from the LTER-MC (Richards et al., 2015), and in a recent exploration of chytrid diversity during blooms of dinoflagellates in the NW Mediterranean Sea (Fernandez-Valero et al., 2022), reveals that the species is common and abundant in the study area, and that Kappamyces representatives also parasitize diatoms in marine waters.
As previously discussed, established marine chytrid-diatom systems are scarce, including Skeletonema (Garvetto et al., 2019), Pseudo-nitzschia pungens (Hanic et al., 2009), benthic diatoms like Navicula spp. and Diploneis didyma (Scholz et al., 2014), or Thalassiosira nordenskioeldii and Chaetoceros sp. (Gaertner, 1979). Although chytrid infections were not observed in natural samples, the Kappamyces representative was able to infect Chaetoceros affinis and Ditylum brightwellii in laboratory incubations, the latter being for the first time observed to be susceptible to chytrid infections. The specificity of the Kappamyces representative cannot be established with the information available in this study, but results obtained from laboratory and co-occurrence analysis suggest it would be able to infect different species in nature.
Numerous statistically significant chytrid-diatom co-occurrence relationships were recorded in the time series, suggesting that many of those chytrid species correspond to diatom parasites. Actually, different characteristics in the patterns of co-occurrences were found. Some chytrid ASV showed positive relationships with numerous diatom ASVs, suggesting those are generalist species able to infect many different hosts. By contrast, many other chytrid ASVs only showed a positive relationship with a single or a few diatom ASVs, suggesting a species-specific host–parasite relationship. In any case, the limited number of detections for each chytrid ASV and their low abundances would require confirmation. However, the identification and validation of such interactions is generally not straightforward because chytrid are not easily observed in nature, requiring a challenging verification in the laboratory due to difficulties in establishing pairwise co-cultures, which impairs a quantitative analysis of the host-parasitoid biological mechanisms. Additionally, and as previously discussed, the detection and characterization of the chytrid community and of their relationships with potential hosts could be hampered by their low abundance compared to other more abundant community components. In fact, rare taxa are most probably sampled and sequenced randomly. Thus, the absence of detections may not represent the absence of chytrids with certainty. Further approaches, like the use of fungal-specific primers or the increase of sequencing depth, could strengthen the soundness of chytrid information and allow for a clearer identification of host–parasite co-occurrences.
In conclusion, the new connections identified in this study widen our view of the microbial food webs and interspecific relationships, improving our knowledge of the actual diversity of the marine communities of the Gulf of Naples. Our results show that fungal chytrids are a significant and dynamic component of the marine microbial community of the LTER-MC site. They are mainly detected in the upper photic layers, where diatoms are abundant, indicating that their role should not be overlooked wherever diatoms represent a dominant component of the plankton community, and should be better addressed with more appropriate and specific approaches.
ACKNOWLEDGMENTS
We acknowledge A. Passarelli, F. Tramontano, M. Cannavacciuolo, and G. Zazo for sampling and the crew of the R/V Vettoria for assistance during the work at sea. We acknowledge the Environmental Monitoring & Analysis (MAA) staff that kindly participated in the laboratory sample processing and chemical analyses. We acknowledge the two anonymous reviewers that helped improving the final version of the manuscript.
FUNDING INFORMATION
The research leading to these results received funding from the European Union's Horizon 2020 Research and Innovation Program under grant agreement No 730984, ASSEMBLE Plus Project. AR and EG were funded by the Spanish MICINN Project SMART (PID2020-112978GB-I00), and thank the institutional support of the ‘Severo Ochoa Centre of Excellence’ accreditation (CEX2019–000928-S). The research program LTER-MC is funded by the Stazione Zoologica Anton Dohrn.
CONFLICT OF INTEREST
All authors state they have no conflicts of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
Data generated in this study including environmental parameters, phytoplankton counts, metabarcoding final datasets and results, images and videos generated from incubation experiments are deposited in the public repository DIGITAL.CSIC, accessible by the DOI 10.20350/digitalCSIC/14744.




